Attaching package: 'pwrss'The following object is masked from 'package:stats':
power.t.testThe pwrss package provides flexible functions for calculating statistical power, required sample size, or detectable effect sizes across a wide range of models, including t-tests, ANOVA, correlation, regression, and generalized linear models. It is designed to handle complex scenarios, such as adjusting for covariates or specifying model-specific parameters like odds ratios or variance explained. This makes it a valuable tool for researchers planning studies to ensure sufficient power and precision in hypothesis testing.
These functions compute statistical power and can plot Type I and II errors if test statistics and degrees of freedom are known.
They’re useful because z, t, $\chi^{2}$, and F stats with degrees of freedom are often reported in publications or software outputs.
Power can be calculated from test statistics as noncentrality parameters, but post-hoc power estimates should be interpreted cautiously.
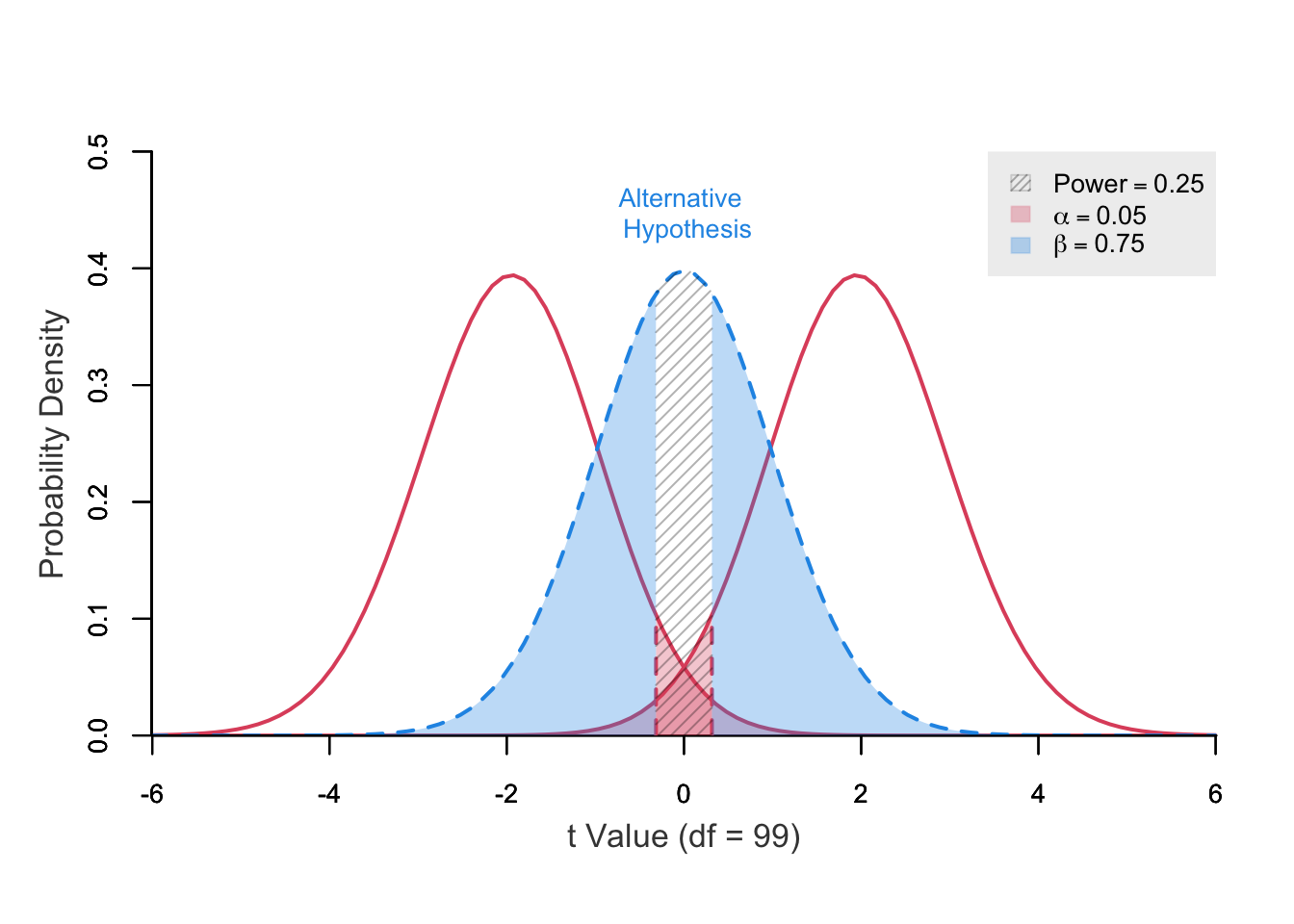
power ncp.alt ncp.null.1 ncp.null.2 alpha df t.crit.1 t.crit.2
0.2371389 0 -1.96 1.96 0.05 99 -0.3155295 0.3155295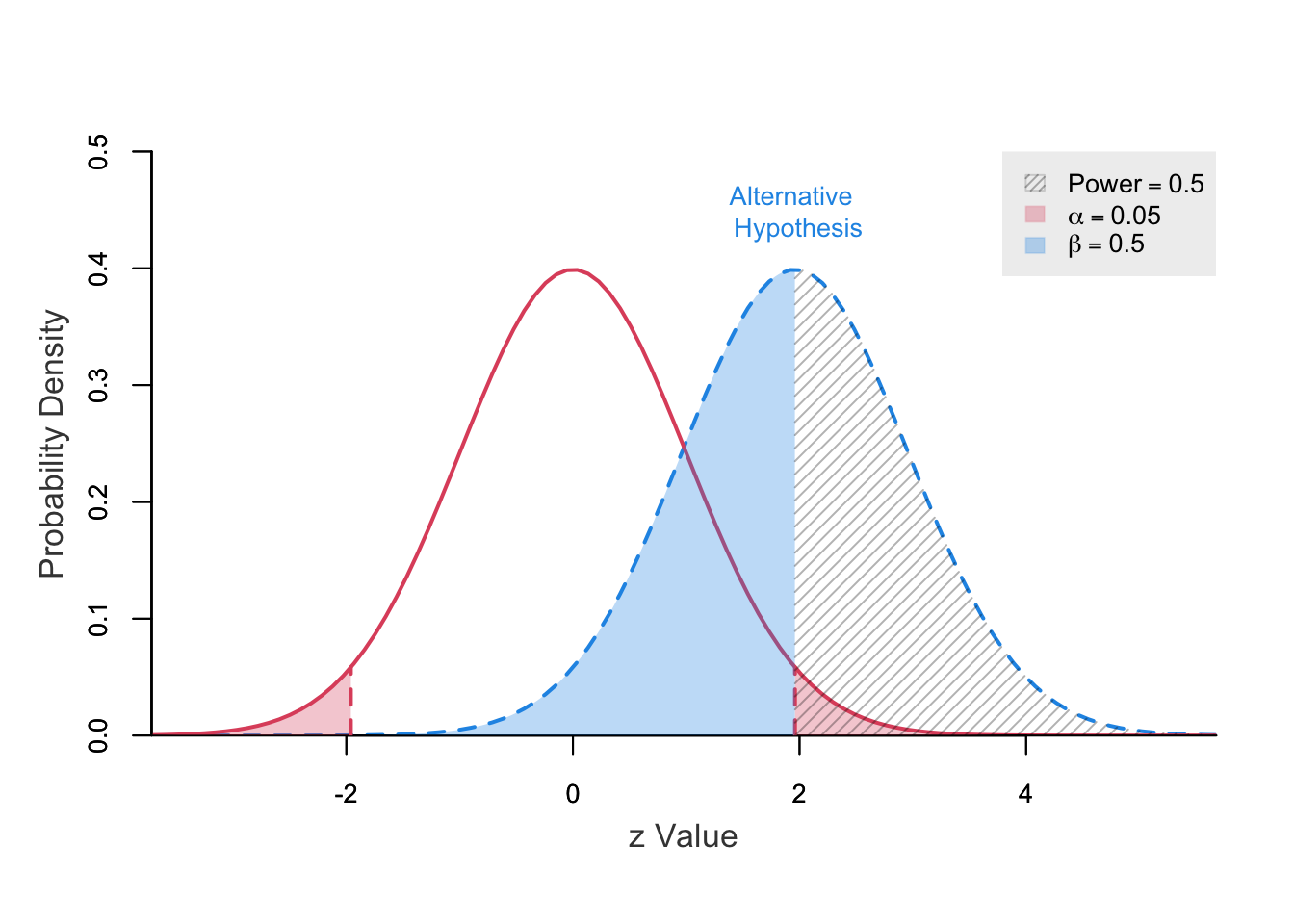
power ncp.alt ncp.null alpha z.crit.1 z.crit.2
0.5000586 1.96 0 0.05 -1.959964 1.959964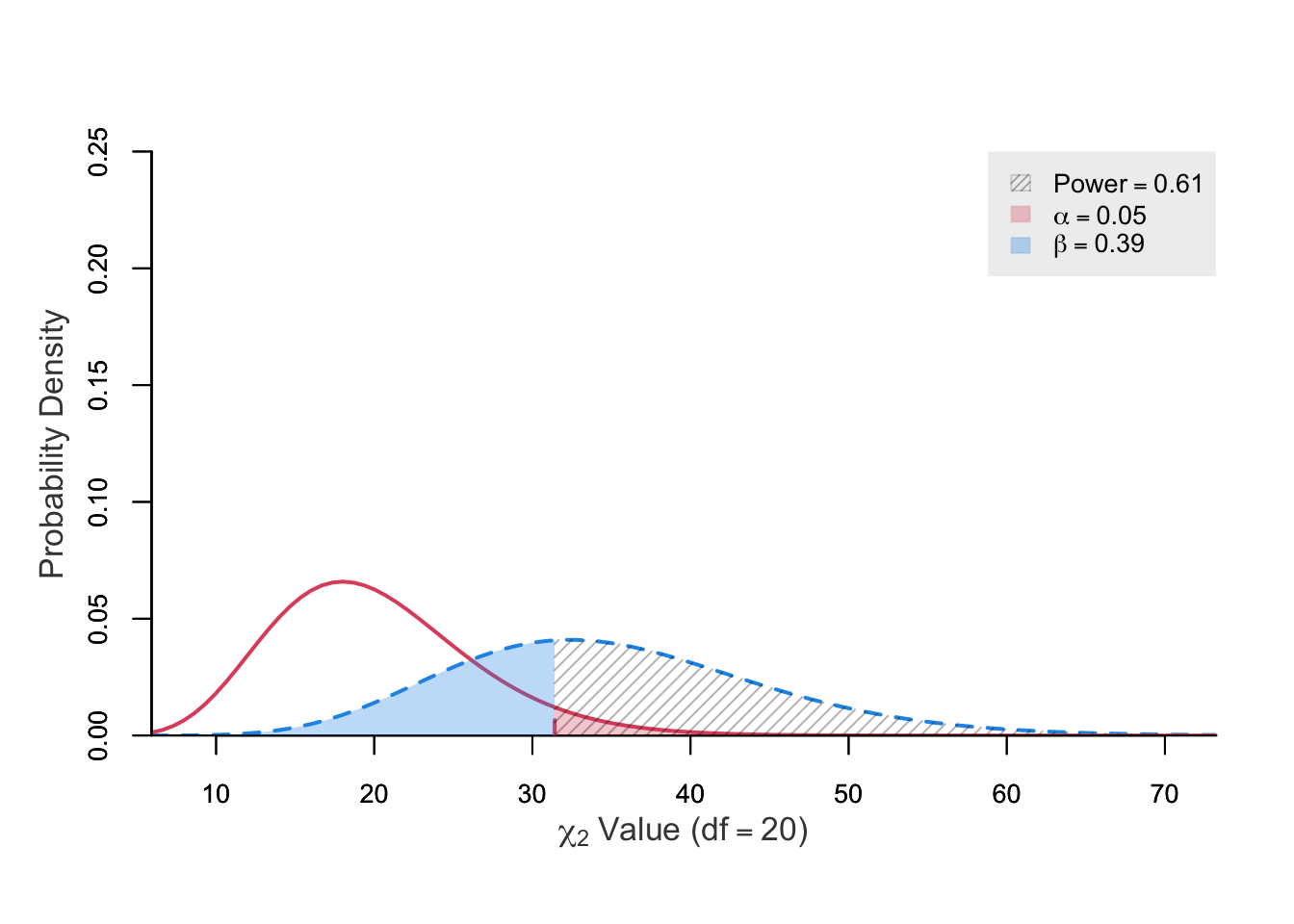
power ncp.alt ncp.null alpha df chisq.crit
0.6110368 15 0 0.05 20 31.41043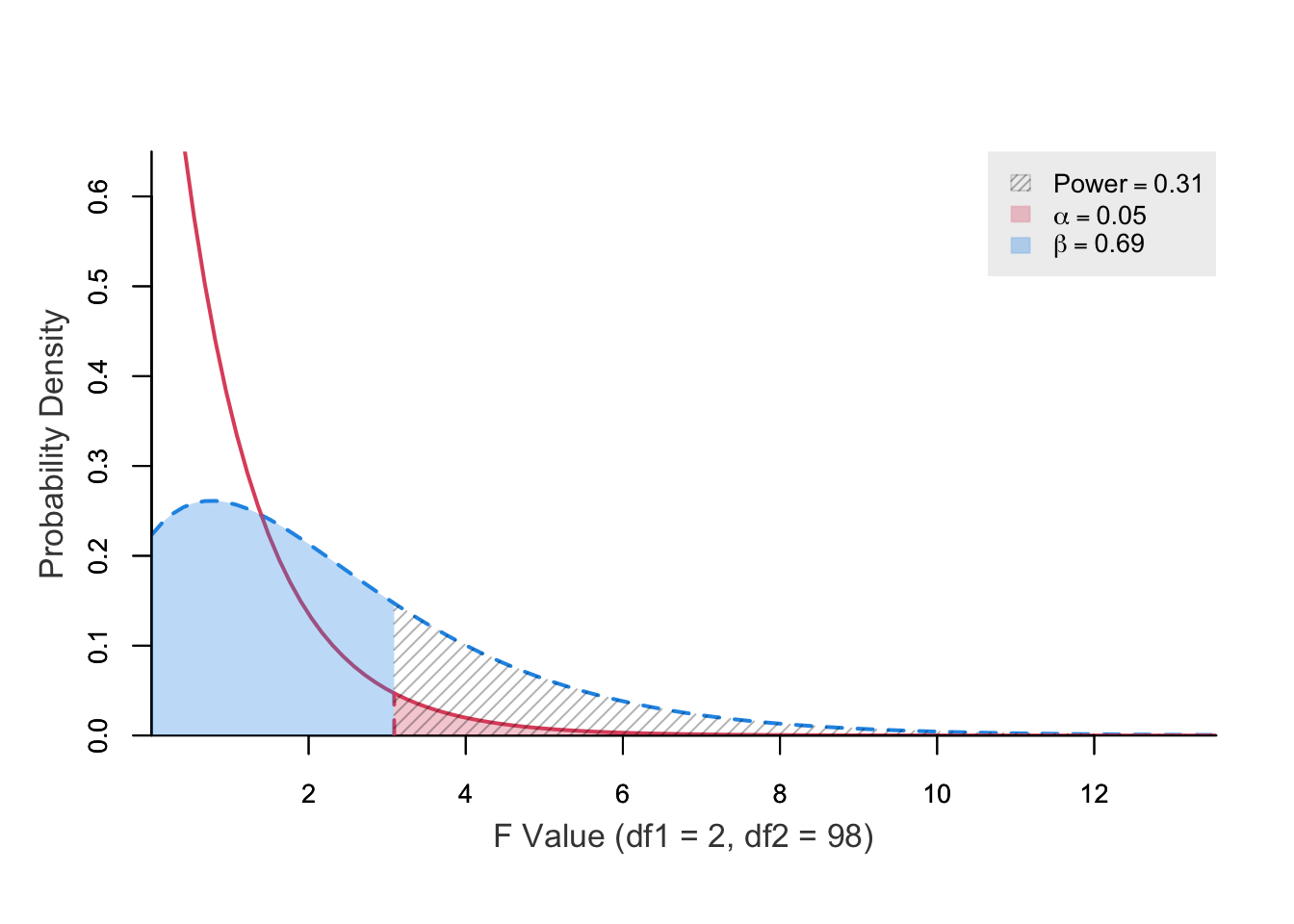
power ncp.alt ncp.null alpha df1 df2 f.crit
0.3128778 3 0 0.05 2 98 3.089203Multiple parameters are allowed but plots should be turned off (plot = FALSE).
power ncp.alt ncp.null alpha df t.crit.1 t.crit.2
0.07852973 0.5 0 0.05 99 -1.984217 1.984217
0.16769955 1.0 0 0.05 99 -1.984217 1.984217
0.31785490 1.5 0 0.05 99 -1.984217 1.984217
0.50826481 2.0 0 0.05 99 -1.984217 1.984217
0.69698027 2.5 0 0.05 99 -1.984217 1.984217 power ncp.alt ncp.null alpha z.crit
0.1291892 1.96 0 0.001 3.090232
0.3570528 1.96 0 0.010 2.326348
0.5000144 1.96 0 0.025 1.959964
0.6236747 1.96 0 0.050 1.644854 power ncp.alt ncp.null alpha df chisq.crit
0.06989507 2 0 0.05 80 101.8795
0.06856779 2 0 0.05 90 113.1453
0.06746196 2 0 0.05 100 124.3421
0.06571411 2 0 0.05 120 146.5674
0.06382959 2 0 0.05 150 179.5806
0.06175379 2 0 0.05 200 233.9943We use the plot() function (S3 method) is a wrapper around the generic functions above. Assign results of any pwrss function to an R object and pass it to plot() function.
Difference between Two means
(Paired Samples t Test)
H0: mu1 - mu2 <= margin
HA: mu1 - mu2 > margin
------------------------------
Statistical power = 0.8
n = 101
------------------------------
Alternative = "non-inferior"
Degrees of freedom = 100
Non-centrality parameter = 2.512
Type I error rate = 0.05
Type II error rate = 0.2 Two-way Analysis of Variance (ANOVA)
H0: 'eta2' or 'f2' = 0
HA: 'eta2' or 'f2' > 0
--------------------------------------
Factor A: 2 levels
Factor B: 3 levels
--------------------------------------
effect power n.total ncp df1 df2
A 0.8 73 8.081 1 66.729
B 0.8 90 9.987 2 83.885
A x B 0.8 90 9.987 2 83.885
--------------------------------------
Type I error rate: 0.05More often than not, unstandardized means and standard deviations are reported in publications for descriptive purposes. Another reason is that they are more intuitive and interpretable (e.g. depression scale). Assume that for the first and second groups expected means are 30 and 28, and expected standard deviations are 12 and 8, respectively.
What is the statistical power given that the sample size for the second group is 50 (n2 = 50) and groups have equal sample sizes (kappa = n1 / n2 = 1)?
Difference between Two means
(Independent Samples t Test)
H0: mu1 = mu2
HA: mu1 != mu2
------------------------------
Statistical power = 0.163
n1 = 50
n2 = 50
------------------------------
Alternative = "not equal"
Degrees of freedom = 98
Non-centrality parameter = 0.981
Type I error rate = 0.05
Type II error rate = 0.837 What is the minimum required sample size given that groups have equal sample sizes (kappa = 1)? (\(\kappa = n_1 / n_2\))
Difference between Two means
(Independent Samples t Test)
H0: mu1 = mu2
HA: mu1 != mu2
------------------------------
Statistical power = 0.8
n1 = 410
n2 = 410
------------------------------
Alternative = "not equal"
Degrees of freedom = 818
Non-centrality parameter = 2.808
Type I error rate = 0.05
Type II error rate = 0.2 It is sufficient to put pooled standard deviation for sd1 because sd2 = sd1 by default. In this case, for a pooled standard deviation of 10.198 the minimum required sample size can be calculated as
Difference between Two means
(Independent Samples t Test)
H0: mu1 = mu2
HA: mu1 != mu2
------------------------------
Statistical power = 0.8
n1 = 410
n2 = 410
------------------------------
Alternative = "not equal"
Degrees of freedom = 818
Non-centrality parameter = 2.808
Type I error rate = 0.05
Type II error rate = 0.2 It is sufficient to put Cohen’s d or Hedge’s g (standardized difference between two groups) for mu1 because mu2 = 0, sd1 = 1, and sd2 = sd1 by default. For example, for an effect size as small as 0.196 (based on previous example) the minimum required sample size can be calculated as
Difference between Two means
(Independent Samples t Test)
H0: mu1 = mu2
HA: mu1 != mu2
------------------------------
Statistical power = 0.8
n1 = 410
n2 = 410
------------------------------
Alternative = "not equal"
Degrees of freedom = 818
Non-centrality parameter = 2.806
Type I error rate = 0.05
Type II error rate = 0.2 Assume for the first (e.g. pretest) and second (e.g. posttest) time points expected means are 30 and 28 (a reduction of 2 points), and expected standard deviations are 12 and 8, respectively. Also assume a correlation of 0.50 between first and second measurements (by default paired.r = 0.50). What is the minimum required sample size?
Difference between Two means
(Paired Samples t Test)
H0: mu1 = mu2
HA: mu1 != mu2
------------------------------
Statistical power = 0.8
n = 222
------------------------------
Alternative = "not equal"
Degrees of freedom = 221
Non-centrality parameter = 2.816
Type I error rate = 0.05
Type II error rate = 0.2 It is sufficient to put standard deviation of the difference for sd1 because sd2 = sd1 by default. In this case, for a standard deviation of difference of 10.583 the minimum required sample size can be calculated as
Difference between Two means
(Paired Samples t Test)
H0: mu1 = mu2
HA: mu1 != mu2
------------------------------
Statistical power = 0.8
n = 222
------------------------------
Alternative = "not equal"
Degrees of freedom = 221
Non-centrality parameter = 2.816
Type I error rate = 0.05
Type II error rate = 0.2 It is sufficient to put Cohen’s d or Hedge’s g (standardized difference between two time points) for mu1 because mu2 = 0, sd1 = sqrt(1/(2*(1-paired.r))), and sd2 = sd1 by default. For example, for an effect size as small as 0.1883 (based on previous example) the minimum required sample size can be calculated as
Difference between Two means
(Paired Samples t Test)
H0: mu1 = mu2
HA: mu1 != mu2
------------------------------
Statistical power = 0.8
n = 224
------------------------------
Alternative = "not equal"
Degrees of freedom = 223
Non-centrality parameter = 2.818
Type I error rate = 0.05
Type II error rate = 0.2 Means might be compared for variables that aren’t normally distributed, due to small samples or inherently non-normal population distributions (e.g. uniform, exponential).
In such cases, t-tests may give biased results.
For non-parametric tests, use pwrss.np.2groups() instead of pwrss.t.2means(), with the same arguments.
"normal", "uniform", "exponential") using the dist argument.Although the function uses means and standard deviations as inputs, it actually tests differences in mean ranks.
Mean differences are converted to Cohen’s d and then to probability of superiority, making it easier to compare and switch between parametric and non-parametric tests.
The example below uses the same parameters as the example in the independent t test section.
Non-parametric Difference between Two Groups (Independent samples)
Mann-Whitney U or Wilcoxon Rank-sum Test
(a.k.a Wilcoxon-Mann-Whitney Test)
Method: GUENTHER
------------------------------
Statistical power = 0.8
n1 = 429
n2 = 429
------------------------------
Alternative = "not equal"
Non-centrality parameter = 2.805
Degrees of freedom = 816.21
Type I error rate = 0.05
Type II error rate = 0.2 The example below uses the same parameters as the example in the paired (dependent) t test section.
It is sufficient to put Cohen’s d or Hedge’s g (standardized difference between two groups or measurements) for mu1 without specifying mu2, sd1, and sd2.
Non-parametric Difference between Two Groups (Dependent samples)
Wilcoxon signed-rank Test for Matched Pairs
Method: GUENTHER
------------------------------
Statistical power = 0.8
n = 233
------------------------------
Alternative = "not equal"
Non-centrality parameter = 2.814
Degrees of freedom = 220.7
Type I error rate = 0.05
Type II error rate = 0.2 These tests are useful for testing practically significant difference (non-inferiority/superiority) or practically null difference (equivalence).
Non-inferiority: The mean of group 1 is practically not smaller than the mean of group 2. The mu1 - mu2 difference can be as small as -1 (margin = -1) but it will still be considered non-inferior. What is the minimum required sample size?
When higher values of an outcome is better the margin takes NEGATIVE values; whereas when lower values of the outcome is better margin takes POSITIVE values.
Difference between Two means
(Independent Samples t Test)
H0: mu1 - mu2 <= margin
HA: mu1 - mu2 > margin
------------------------------
Statistical power = 0.8
n1 = 144
n2 = 144
------------------------------
Alternative = "non-inferior"
Degrees of freedom = 286
Non-centrality parameter = 2.496
Type I error rate = 0.05
Type II error rate = 0.2 Superiority: The mean of group 1 is practically greater than the mean of group 2. The mu1 - mu2 difference is at least greater than 1 (margin = 1). What is the minimum required sample size?
When higher values of an outcome is better margin takes POSITIVE values; whereas when lower values of the outcome is better margin takes NEGATIVE values.
Difference between Two means
(Independent Samples t Test)
H0: mu1 - mu2 <= margin
HA: mu1 - mu2 > margin
------------------------------
Statistical power = 0.8
n1 = 1287
n2 = 1287
------------------------------
Alternative = "superior"
Degrees of freedom = 2572
Non-centrality parameter = 2.487
Type I error rate = 0.05
Type II error rate = 0.2 Equivalence: The mean of group 1 is practically same as mean of group 2. The mu1 - mu2 difference can be as small as -1 and as high as 1 (margin = 1). What is the minimum required sample size?
Specify the absolute value for the margin.
Difference between Two means
(Independent Samples t Test)
H0: |mu1 - mu2| >= margin
HA: |mu1 - mu2| < margin
------------------------------
Statistical power = 0.8
n1 = 1783
n2 = 1783
------------------------------
Alternative = "equivalent"
Degrees of freedom = 3564
Non-centrality parameter = -2.928
Type I error rate = 0.05
Type II error rate = 0.2 Non-inferiority: The mean of group 1 is practically not smaller than the mean of group 2. The mu1 - mu2 difference can be as small as -1 (margin = -1). What is the minimum required sample size?
Non-parametric Difference between Two Groups (Independent samples)
Mann-Whitney U or Wilcoxon Rank-sum Test
(a.k.a Wilcoxon-Mann-Whitney Test)
Method: GUENTHER
------------------------------
Statistical power = 0.8
n1 = 151
n2 = 151
------------------------------
Alternative = "non-inferior"
Non-centrality parameter = 2.492
Degrees of freedom = 285.13
Type I error rate = 0.05
Type II error rate = 0.2 Superiority: The mean of group 1 is practically greater than the mean of group 2. The mu1 - mu2 difference is at least greater than 1 (margin = 1). What is the minimum required sample size?
Non-parametric Difference between Two Groups (Independent samples)
Mann-Whitney U or Wilcoxon Rank-sum Test
(a.k.a Wilcoxon-Mann-Whitney Test)
Method: GUENTHER
------------------------------
Statistical power = 0.8
n1 = 1348
n2 = 1348
------------------------------
Alternative = "superior"
Non-centrality parameter = 2.487
Degrees of freedom = 2571.3
Type I error rate = 0.05
Type II error rate = 0.2 Equivalence: The mean of group 1 is practically same as mean of group 2. The mu1 - mu2 difference can be as small as -1 and as high as 1 (margin = 1). What is the minimum required sample size?
Non-parametric Difference between Two Groups (Independent samples)
Mann-Whitney U or Wilcoxon Rank-sum Test
(a.k.a Wilcoxon-Mann-Whitney Test)
Method: GUENTHER
------------------------------
Statistical power = 0.8
n1 = 1867
n2 = 1867
------------------------------
Alternative = "equivalent"
Non-centrality parameter = -2.927
Degrees of freedom = 3561.91
Type I error rate = 0.05
Type II error rate = 0.2 Omnibus F test in multiple liner regression is used to test whether \(R^2\) is greater than 0 (zero). Assume that we want to predict a continuous variable \(Y\) using \(X_{1}\), \(X_{2}\), and \(X_{2}\) variables (a combination of binary or continuous).
\[\begin{eqnarray} Y &=& \beta_{0} + \beta_{1}X_{1} + \beta_{2}X_{2} + \beta_{3}X_{3} + r, \quad r \thicksim N(0,\sigma^2) \newline \end{eqnarray}\]
We are expecting that these three variables explain 30% of the variance in the outcome (\(R^2 = 0.30\) or r2 = 0.30 in the code). What is the minimum required sample size?
Linear Regression (F test)
R-squared Deviation from 0 (zero)
H0: r2 = 0
HA: r2 > 0
------------------------------
Statistical power = 0.8
n = 30
------------------------------
Numerator degrees of freedom = 3
Denominator degrees of freedom = 25.653
Non-centrality parameter = 12.709
Type I error rate = 0.05
Type II error rate = 0.2 In the earlier example, assume that we want to predict a continuous variable \(Y\) using a continuous predictor \(X_{1}\) but control for \(X_{2}\), and \(X_{2}\) variables (a combination of binary or continuous). We are mainly interested in the effect of \(X_{1}\) and expect a standardized regression coefficient of \(\beta_{1} = 0.20\).
\[\begin{eqnarray} Y &=& \beta_{0} + \color{red} {\beta_{1} X_{1}} + \beta_{2}X_{2} + \beta_{3}X_{3} + r, \quad r \thicksim N(0,\sigma^2) \newline \end{eqnarray}\]
Again, we are expecting that these three variables explain 30% of the variance in the outcome (\(R^2 = 0.30\)). What is the minimum required sample size? It is sufficient to provide standardized regression coefficient for beta1 because sdx = 1 and sdy = 1 by default.
Linear Regression Coefficient (t Test)
H0: beta1 = beta0
HA: beta1 != beta0
------------------------------
Statistical power = 0.8
n = 140
------------------------------
Alternative = "not equal"
Degrees of freedom = 135.331
Non-centrality parameter = 2.822
Type I error rate = 0.05
Type II error rate = 0.2 For unstandardized coefficients specify sdy and sdx. Assume we are expecting an unstandardized regression coefficient of beta1 = 0.60, a standard deviation of sdy = 12 for the outcome and a standard deviation of sdx = 4 for the main predictor. What is the minimum required sample size?
Linear Regression Coefficient (t Test)
H0: beta1 = beta0
HA: beta1 != beta0
------------------------------
Statistical power = 0.8
n = 140
------------------------------
Alternative = "not equal"
Degrees of freedom = 135.331
Non-centrality parameter = 2.822
Type I error rate = 0.05
Type II error rate = 0.2 When the main predictor is binary (e.g. treatment/control), its standardized regression coefficient equals Cohen’s d.
The predictor’s standard deviation is $\sqrt{p(1-p)}$, where $p$ is the group proportion; for $p = 0.50$, what’s the minimum required sample size?
Provide Cohen’s d for β₁ and set sdx = sqrt(p*(1-p)) when calculating.
Linear Regression Coefficient (t Test)
H0: beta1 = beta0
HA: beta1 != beta0
------------------------------
Statistical power = 0.8
n = 552
------------------------------
Alternative = "not equal"
Degrees of freedom = 547.355
Non-centrality parameter = 2.807
Type I error rate = 0.05
Type II error rate = 0.2 These tests are useful for testing practically significant effects (non-inferiority/superiority) or practically null effects (equivalence).
The intervention is expected to be non-inferior to some earlier or other interventions. Assume that the effect of an earlier or some other intervention is beta0 = 0.10. The beta1 - beta0 is expected to be positive and should be at least -0.05 (margin = -0.05). What is the minimum required sample size?
This is the case when higher values of an outcome is better. When lower values of an outcome is better the beta1 - beta0 difference is expected to be NEGATIVE and the margin takes POSITIVE values.
Linear Regression Coefficient (t Test)
H0: beta1 - beta0 <= margin
HA: beta1 - beta0 > margin
------------------------------
Statistical power = 0.8
n = 771
------------------------------
Alternative = "non-inferior"
Degrees of freedom = 766.745
Non-centrality parameter = 2.489
Type I error rate = 0.05
Type II error rate = 0.2 The intervention is expected to be superior to some earlier or other interventions. Assume that the effect of an earlier or some other intervention is beta0 = 0.10. The beta1 - beta0 is expected to be positive and should be at least 0.05 (margin = 0.05). What is the minimum required sample size?
This is the case when higher values of an outcome is better. When lower values of an outcome is better beta1 - beta0 difference is expected to be NEGATIVE and the margin takes NEGATIVE values.
Linear Regression Coefficient (t Test)
H0: beta1 - beta0 <= margin
HA: beta1 - beta0 > margin
------------------------------
Statistical power = 0.8
n = 6926
------------------------------
Alternative = "superior"
Degrees of freedom = 6921.818
Non-centrality parameter = 2.487
Type I error rate = 0.05
Type II error rate = 0.2 The intervention is expected to be equivalent to some earlier or other interventions. Assume the effect of an earlier or some other intervention is beta0 = 0.20. The beta1 - beta0 is expected to be within -0.05 and 0.05 (margin = 0.05). What is the minimum required sample size?
margin always takes positive values for equivalence. Specify the absolute value.
Linear Regression Coefficient (t Test)
H0: |beta1 - beta0| >= margin
HA: |beta1 - beta0| < margin
------------------------------
Statistical power = 0.8
n = 9593
------------------------------
Alternative = "equivalent"
Degrees of freedom = 9588.862
Non-centrality parameter = -2.927 2.927
Type I error rate = 0.05
Type II error rate = 0.2 In logistic regression a binary outcome variable (0/1: failed/passed, dead/alive, absent/present) is modeled by predicting probability of being in group 1 (\(P_1\)) via logit transformation (natural logarithm of odds). The base probability \(P_0\) is the overall probability of being in group 1 without influence of predictors in the model (null). Under alternative hypothesis, the probability of being in group 1 (\(P_1\)) deviate from \(P_0\) depending on the value of the predictor; whereas under null it is same as the \(P_0\).
A model with one main predictor (\(X_1\)) and two other covariates (\(X_2\) and \(X_3\)) can be constructed as
\[\begin{eqnarray} ln(\frac{P_1}{1- P_1}) &=& \beta_{0} + \color{red} {\beta_{1} X_{1}} + \beta_{2}X_{2} + \beta_{3}X_{3} \newline \end{eqnarray}\]
Therefore the odds ratio is defined as \[OR = exp(\beta_1) = \frac{P_1}{1- P_1} / \frac{P_0}{1- P_0}\]
r2.other.x = 0.20 in the code). It can be found in the form of adjusted R-square via regressing \(X_1\) on \(X_2\) and \(X_3\). Higher values require larger sample sizes. The default is 0 (zero).What is the minimum required sample size? There are three types of specification to statistical power or sample size calculations; (i) probability specification, (ii) odds ratio specification, and (iii) regression coefficient specification (as in standard software output).
Probability specification:
Logistic Regression Coefficient
(Large Sample Approx. Wald's z Test)
H0: beta1 = 0
HA: beta1 != 0
Distribution of X = 'normal'
Method = DEMIDENKO(VC)
------------------------------
Statistical power = 0.8
n = 365
------------------------------
Alternative = "not equal"
Non-centrality parameter = -2.766
Type I error rate = 0.05
Type II error rate = 0.2 Odds ratio specification: \[OR = \frac{P_1}{1-P1} / \frac{P_0}{1-P_0} = \frac{0.10}{1-0.10} / \frac{0.15}{1-0.15} = 0.6296\]
Logistic Regression Coefficient
(Large Sample Approx. Wald's z Test)
H0: beta1 = 0
HA: beta1 != 0
Distribution of X = 'normal'
Method = DEMIDENKO(VC)
------------------------------
Statistical power = 0.8
n = 365
------------------------------
Alternative = "not equal"
Non-centrality parameter = -2.766
Type I error rate = 0.05
Type II error rate = 0.2 Regression coefficient specification: \[\beta_1 = ln(\frac{P_1}{1-P1} / \frac{P_0}{1-P_0}) = ln(0.6296) = -0.4626\]
Logistic Regression Coefficient
(Large Sample Approx. Wald's z Test)
H0: beta1 = 0
HA: beta1 != 0
Distribution of X = 'normal'
Method = DEMIDENKO(VC)
------------------------------
Statistical power = 0.8
n = 365
------------------------------
Alternative = "not equal"
Non-centrality parameter = -2.766
Type I error rate = 0.05
Type II error rate = 0.2 Change the distribution’s parameters for predictor X:
The mean and standard deviation of a normally distributed main predictor is 0 and 1 by default. They can be modified. In the following example the mean is 25 and the standard deviation is 8.
Logistic Regression Coefficient
(Large Sample Approx. Wald's z Test)
H0: beta1 = 0
HA: beta1 != 0
Distribution of X = 'normal'
Method = DEMIDENKO(VC)
------------------------------
Statistical power = 0.8
n = 2435
------------------------------
Alternative = "not equal"
Non-centrality parameter = -2.423
Type I error rate = 0.05
Type II error rate = 0.2 Change the distribution family of predictor X:
More distribution types are supported by the function. For example, the main predictor can be binary (e.g. treatment/control groups). Often half of the sample is assigned to the treatment group and the other half to the control (prob = 0.50 by default).
Logistic Regression Coefficient
(Large Sample Approx. Wald's z Test)
H0: beta1 = 0
HA: beta1 != 0
Distribution of X = 'bernoulli'
Method = DEMIDENKO(VC)
------------------------------
Statistical power = 0.8
n = 1723
------------------------------
Alternative = "not equal"
Non-centrality parameter = -2.789
Type I error rate = 0.05
Type II error rate = 0.2 Change the treatment group allocation rate of the binary predictor X (prob = 0.40):
Sometimes treatment groups cost more per subject or are harder to recruit than control groups, making an unbalanced sample practical. For example, with 40% of subjects in the treatment group, what is the minimum required sample size?
Logistic Regression Coefficient
(Large Sample Approx. Wald's z Test)
H0: beta1 = 0
HA: beta1 != 0
Distribution of X = 'bernoulli'
Method = DEMIDENKO(VC)
------------------------------
Statistical power = 0.8
n = 1826
------------------------------
Alternative = "not equal"
Non-centrality parameter = -2.766
Type I error rate = 0.05
Type II error rate = 0.2 One-way ANOVA/ANCOVA: Compares means across groups; ANCOVA adjusts for covariates.
Two- or Three-way ANOVA/ANCOVA: Examines interactions between factors; ANCOVA adjusts interactions for covariates and determines required sample size for complex designs.
A researcher is expecting a difference of Cohen’s d = 0.50 between treatment and control groups (two levels) translating into \(\eta^2 = 0.059\) (eta2 = 0.059). Means are not adjusted for any covariates. What is the minimum required sample size?
One-way Analysis of Variance (ANOVA)
H0: 'eta2' or 'f2' = 0
HA: 'eta2' or 'f2' > 0
--------------------------------------
Factor A: 2 levels
--------------------------------------
effect power n.total ncp df1 df2
A 0.8 128 7.971 1 125.132
--------------------------------------
Type I error rate: 0.05A researcher is expecting an adjusted difference of Cohen’s d = 0.45 between treatment and control groups (n.levels = 2) after controlling for the pretest (n.cov = 1) translating into partial \(\eta^2 = 0.048\) (eta2 = 0.048). What is the minimum required sample size?
n.cov (or n.covariates) argument has trivial effect on the results. The difference between ANOVA and ANCOVA procedure depends on whether the effect (eta2) is unadjusted or covariate-adjusted.
One-way Analysis of Covariance (ANCOVA)
H0: 'eta2' or 'f2' = 0
HA: 'eta2' or 'f2' > 0
--------------------------------------
Factor A: 2 levels
--------------------------------------
effect power n.total ncp df1 df2
A 0.8 158 7.948 1 154.626
--------------------------------------
Type I error rate: 0.05A researcher is expecting a partial \(\eta^2 = 0.03\) (eta2 = 0.03) for interaction of treatment/control (Factor A: two levels) with gender (Factor B: two levels). Thus, n.levels = c(2,2). What is the minimum required sample size?
Two-way Analysis of Variance (ANOVA)
H0: 'eta2' or 'f2' = 0
HA: 'eta2' or 'f2' > 0
--------------------------------------
Factor A: 2 levels
Factor B: 2 levels
--------------------------------------
effect power n.total ncp df1 df2
A 0.8 256 7.909 1 251.73
B 0.8 256 7.909 1 251.73
A x B 0.8 256 7.909 1 251.73
--------------------------------------
Type I error rate: 0.05A researcher is expecting a partial \(\eta^2 = 0.02\) (eta2 = 0.02) for interaction of treatment/control (Factor A) with gender (Factor B) adjusted for the pretest (n.cov = 1). What is the minimum required sample size?
Two-way Analysis of Covariance (ANCOVA)
H0: 'eta2' or 'f2' = 0
HA: 'eta2' or 'f2' > 0
--------------------------------------
Factor A: 2 levels
Factor B: 2 levels
--------------------------------------
effect power n.total ncp df1 df2
A 0.8 387 7.889 1 381.539
B 0.8 387 7.889 1 381.539
A x B 0.8 387 7.889 1 381.539
--------------------------------------
Type I error rate: 0.05One-sided Test: Assume that the expected correlation is 0.20 and it is greater than 0.10. What is the minimum required sample size?
A Correlation against a Constant (z Test)
H0: r = r0
HA: r > r0
------------------------------
Statistical power = 0.8
n = 593
------------------------------
Alternative = "greater"
Non-centrality parameter = 2.486
Type I error rate = 0.05
Type II error rate = 0.2 Two-sided Test: Assume that the expected correlation is 0.20 and it is different from 0 (zero). The correlation could be 0.20 as well as -0.20. What is the minimum required sample size?
A Correlation against a Constant (z Test)
H0: r = r0
HA: r != r0
------------------------------
Statistical power = 0.8
n = 194
------------------------------
Alternative = "not equal"
Non-centrality parameter = 2.802
Type I error rate = 0.05
Type II error rate = 0.2 Assume that the expected correlations in the first and second groups are 0.30 and 0.20, respectively (r1 = 0.30 and r2 = 0.20).
One-sided Test: Expecting r1 - r2 greater than 0 (zero). The difference could be 0.10 but could not be -0.10. What is the minimum required sample size?
Difference between Two Correlations
(Independent Samples z Test)
H0: r1 = r2
HA: r1 > r2
------------------------------
Statistical power = 0.8
n1 = 1088
n2 = 1088
------------------------------
Alternative = "greater"
Non-centrality parameter = 2.486
Type I error rate = 0.05
Type II error rate = 0.2 Two-sided Test: Expecting r1 - r2 different from 0 (zero). The difference could be -0.10 as well as 0.10. What is the minimum required sample size?
Difference between Two Correlations
(Independent Samples z Test)
H0: r1 = r2
HA: r1 != r2
------------------------------
Statistical power = 0.8
n1 = 1380
n2 = 1380
------------------------------
Alternative = "not equal"
Non-centrality parameter = 2.802
Type I error rate = 0.05
Type II error rate = 0.2 In the following examples p is the proportion under alternative hypothesis and p0 is the proportion under null hypothesis.
One-sided Test: Expecting p - p0 smaller than 0 (zero).
Approach: Normal Approximation
A Proportion against a Constant (z Test)
H0: p = p0
HA: p < p0
------------------------------
Statistical power = 0.8
n = 613
------------------------------
Alternative = "less"
Non-centrality parameter = -2.486
Type I error rate = 0.05
Type II error rate = 0.2 # arcsine transformation
pwrss.z.prop(p = 0.45, p0 = 0.50,
alpha = 0.05, power = 0.80,
alternative = "less",
arcsin.trans = TRUE) Approach: Arcsine Transformation
A Proportion against a Constant (z Test)
H0: p = p0
HA: p < p0
------------------------------
Statistical power = 0.8
n = 617
------------------------------
Alternative = "less"
Non-centrality parameter = -2.486
Type I error rate = 0.05
Type II error rate = 0.2 Two-sided Test: Expecting p - p0 smaller than 0 (zero) or greater than 0 (zero).
Approach: Normal Approximation
A Proportion against a Constant (z Test)
H0: p = p0
HA: p != p0
------------------------------
Statistical power = 0.8
n = 778
------------------------------
Alternative = "not equal"
Non-centrality parameter = -2.802
Type I error rate = 0.05
Type II error rate = 0.2 Non-inferiority Test: The case when smaller proportion is better. Expecting p - p0 smaller than 0.01.
Approach: Normal Approximation
A Proportion against a Constant (z Test)
H0: p - p0 <= margin
HA: p - p0 > margin
------------------------------
Statistical power = 0.8
n = 426
------------------------------
Alternative = "non-inferior"
Non-centrality parameter = -2.486
Type I error rate = 0.05
Type II error rate = 0.2 Non-inferiority Test: The case when bigger proportion is better. Expecting p - p0 greater than -0.01.
Approach: Normal Approximation
A Proportion against a Constant (z Test)
H0: p - p0 <= margin
HA: p - p0 > margin
------------------------------
Statistical power = 0.8
n = 426
------------------------------
Alternative = "non-inferior"
Non-centrality parameter = 2.486
Type I error rate = 0.05
Type II error rate = 0.2 Superiority Test: The case when smaller proportion is better. Expecting p - p0 smaller than -0.01.
Approach: Normal Approximation
A Proportion against a Constant (z Test)
H0: p - p0 <= margin
HA: p - p0 > margin
------------------------------
Statistical power = 0.8
n = 957
------------------------------
Alternative = "superior"
Non-centrality parameter = -2.486
Type I error rate = 0.05
Type II error rate = 0.2 Superiority Test: The case when bigger proportion is better. Expecting p - p0 greater than 0.01.
Approach: Normal Approximation
A Proportion against a Constant (z Test)
H0: p - p0 <= margin
HA: p - p0 > margin
------------------------------
Statistical power = 0.8
n = 957
------------------------------
Alternative = "superior"
Non-centrality parameter = 2.486
Type I error rate = 0.05
Type II error rate = 0.2 Equivalence Test: Expecting p - p0 between -0.01 and 0.01.
Approach: Normal Approximation
A Proportion against a Constant (z Test)
H0: |p - p0| >= margin
HA: |p - p0| < margin
------------------------------
Statistical power = 0.8
n = 21410
------------------------------
Alternative = "equivalent"
Non-centrality parameter = -2.926 2.926
Type I error rate = 0.05
Type II error rate = 0.2 In the following examples p1 and p2 are proportions for the first and second groups under alternative hypothesis. The null hypothesis state p1 = p2 or p1 - p2 = 0.
One-sided Test: Expecting p1 - p2 smaller than 0 (zero).
Approach: Normal Approximation
Difference between Two Proportions
(Independent Samples z Test)
H0: p1 = p2
HA: p1 < p2
------------------------------
Statistical power = 0.8
n1 = 1231
n2 = 1231
------------------------------
Alternative = "less"
Non-centrality parameter = -2.486
Type I error rate = 0.05
Type II error rate = 0.2 Two-sided Test: Expecting p1 - p2 smaller than 0 (zero) or greater than 0 (zero).
Approach: Normal Approximation
Difference between Two Proportions
(Independent Samples z Test)
H0: p1 = p2
HA: p1 != p2
------------------------------
Statistical power = 0.8
n1 = 1562
n2 = 1562
------------------------------
Alternative = "not equal"
Non-centrality parameter = -2.802
Type I error rate = 0.05
Type II error rate = 0.2 Non-inferiority Test: The case when smaller proportion is better. Expecting p1 - p2 smaller than 0.01.
Approach: Normal Approximation
Difference between Two Proportions
(Independent Samples z Test)
H0: p1 - p2 <= margin
HA: p1 - p2 > margin
------------------------------
Statistical power = 0.8
n1 = 855
n2 = 855
------------------------------
Alternative = "non-inferior"
Non-centrality parameter = -2.486
Type I error rate = 0.05
Type II error rate = 0.2 Non-inferiority Test: The case when bigger proportion is better. Expecting p1 - p2 greater than -0.01.
Approach: Normal Approximation
Difference between Two Proportions
(Independent Samples z Test)
H0: p1 - p2 <= margin
HA: p1 - p2 > margin
------------------------------
Statistical power = 0.8
n1 = 855
n2 = 855
------------------------------
Alternative = "non-inferior"
Non-centrality parameter = 2.486
Type I error rate = 0.05
Type II error rate = 0.2 Superiority Test: The case when smaller proportion is better. Expecting p1 - p2 smaller than -0.01.
Approach: Normal Approximation
Difference between Two Proportions
(Independent Samples z Test)
H0: p1 - p2 <= margin
HA: p1 - p2 > margin
------------------------------
Statistical power = 0.8
n1 = 1923
n2 = 1923
------------------------------
Alternative = "superior"
Non-centrality parameter = -2.486
Type I error rate = 0.05
Type II error rate = 0.2 Superiority Test: The case when bigger proportion is better. Expecting p1 - p2 greater than 0.01.
Approach: Normal Approximation
Difference between Two Proportions
(Independent Samples z Test)
H0: p1 - p2 <= margin
HA: p1 - p2 > margin
------------------------------
Statistical power = 0.8
n1 = 1923
n2 = 1923
------------------------------
Alternative = "superior"
Non-centrality parameter = 2.486
Type I error rate = 0.05
Type II error rate = 0.2 Equivalence Test: Expecting p1 - p2 between -0.01 and 0.01.
Approach: Normal Approximation
Difference between Two Proportions
(Independent Samples z Test)
H0: |p1 - p2| >= margin
HA: |p1 - p2| < margin
------------------------------
Statistical power = 0.8
n1 = 42820
n2 = 42820
------------------------------
Alternative = "equivalent"
Non-centrality parameter = -2.926 2.926
Type I error rate = 0.05
Type II error rate = 0.2