set.seed(134125)
sim_parms <- list(
n = 110,
alpha = 2.2,
beta = -1.37,
mu_x = 2,
sigma_x = 2,
LoD = 0
)6 Example Model 4: Logistic regression with censored predictors
For the next example model we have enough background knowledge to implement a model for a problem that my group has been trying to tackle recently, and that is somewhat common with the type of data that I work with. Suppose we want to test the efficacy of a novel vaccine candidate in a clinical trial.
Typically, these vaccines have immunological correlates of protection, some value we can measure that gives a general idea of how strong a person’s immune response was, and correlates with their probability of getting a disease. One of the most common types of these measurements are antibody titers, where typically a high antibody titer is protective and corresponds to a lower probability of disease. If we have a clinical trial where we record if patients get the disease (after challenge or after a sufficiently long followup period, for this example the details are not too important as long as we assume the infection status is measured accurately), we can model how these measurements are related to protection via logistic regression.
6.1 Model with one patient group
For the first model, we’ll consider a simpler case where we only have one group of patients. This model would be appropriate for, e.g., an observational study where all patients are given the vaccine of interest.
The data generating model is as follows:
mostly cause it’s annoying and confusing to write out all the logs every time, so we could also write
Now that we have the data generating process written out, we can simulate some example data. Note that in this example, we can interpret
We then want our true
inv_logit <- function(x) {return(1 / (1 + exp(-x)))}
sim_one_group <- function(n, alpha, beta, mu_x, sigma_x, LoD) {
out <- tibble::tibble(
x_star = rnorm(n, mu_x, sigma_x),
x = ifelse(x_star < LoD, 0.5 * LoD, x_star),
p = inv_logit(alpha + beta * x_star),
y = rbinom(n, size = 1, prob = p)
)
}
sim_data <- do.call(sim_one_group, sim_parms)Of course visualizing the relationship between a binary outcome and a continuous predictor is in some sense more complex than visualizing the relationship between a continuous outcome and a continuous predictor.
First, let’s look at how the distribution of the predictor variable changes if we condition on the outcome.
sim_data |>
tidyr::pivot_longer(cols = c(x, x_star)) |>
dplyr::mutate(
yf = factor(
y,
levels = c(0, 1),
labels = c("Not infected", "Infected")
),
name = factor(
name,
levels = c("x_star", "x"),
labels = c("Latent variable", "Observed variable")
)
) |>
ggplot() +
aes(x = value, fill = yf) +
geom_vline(
xintercept = 0,
linetype = "dashed",
color = "black",
linewidth = 1
) +
geom_histogram(
binwidth = 0.5, boundary = 0, closed = "left",
position = "identity", alpha = 0.6,
color = "black"
) +
scale_x_continuous(
name = "Simulated log titer",
breaks = scales::breaks_pretty()
) +
scale_y_continuous(breaks = scales::breaks_pretty()) +
scale_fill_brewer(palette = "Dark2", name = NULL) +
facet_wrap(facets = vars(name))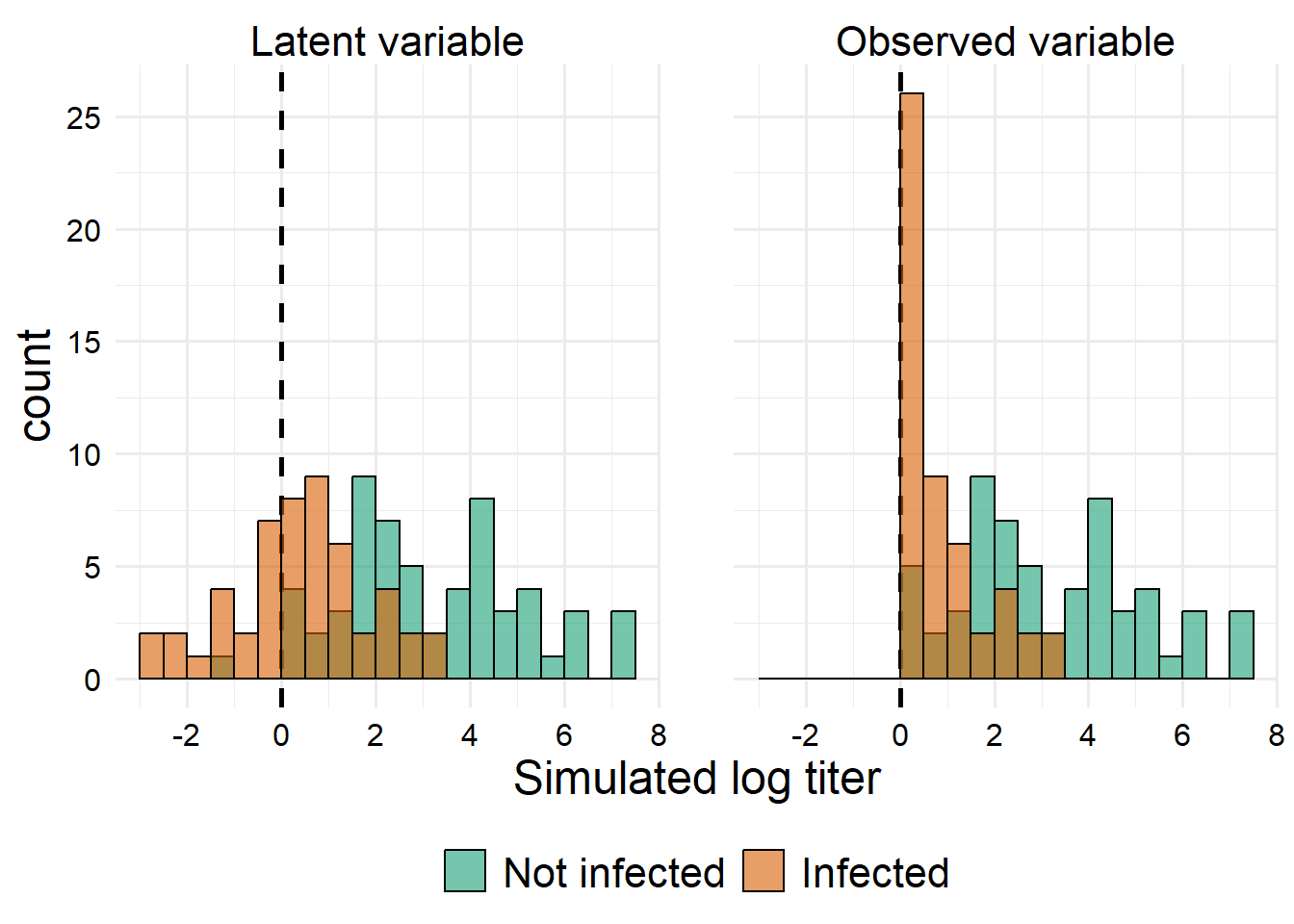
Essentially everything below our lower threshold gets bumped up, which will make the summary statistics of the distribution more similar between groups for the observed titer values than they would have been for the latent titer values. However, we can still see a large difference.
interp <-
tibble::tibble(
value = seq(-2, 6, 0.1),
p = inv_logit(sim_parms$alpha + sim_parms$beta * value)
)
interp2 <-
dplyr::bind_rows(
"Latent variable" = interp,
"Observed variable" = interp,
.id = "name"
)
lab1 <- latex2exp::TeX(r"($Pr(y_{i} = 1 \ | \ x_{i})$)")
sim_data |>
tidyr::pivot_longer(cols = c(x, x_star)) |>
dplyr::mutate(
yf = factor(
y,
levels = c(0, 1),
labels = c("Not infected", "Infected")
),
name = factor(
name,
levels = c("x_star", "x"),
labels = c("Latent variable", "Observed variable")
)
) |>
ggplot() +
aes(x = value, color = name) +
geom_line(
data = interp2, aes(y = p), color = "darkgray", linetype = 2, linewidth = 1
) +
geom_point(
aes(y = y), size = 3, alpha = 0.5,
position = position_jitter(width = 0, height = 0.05, seed = 370)
) +
scale_x_continuous(
name = "Simulated log titer",
breaks = scales::breaks_pretty()
) +
scale_y_continuous(
name = lab1,
breaks = scales::breaks_pretty()
) +
scale_color_brewer(palette = "Accent", name = NULL) +
facet_wrap(facets = vars(name))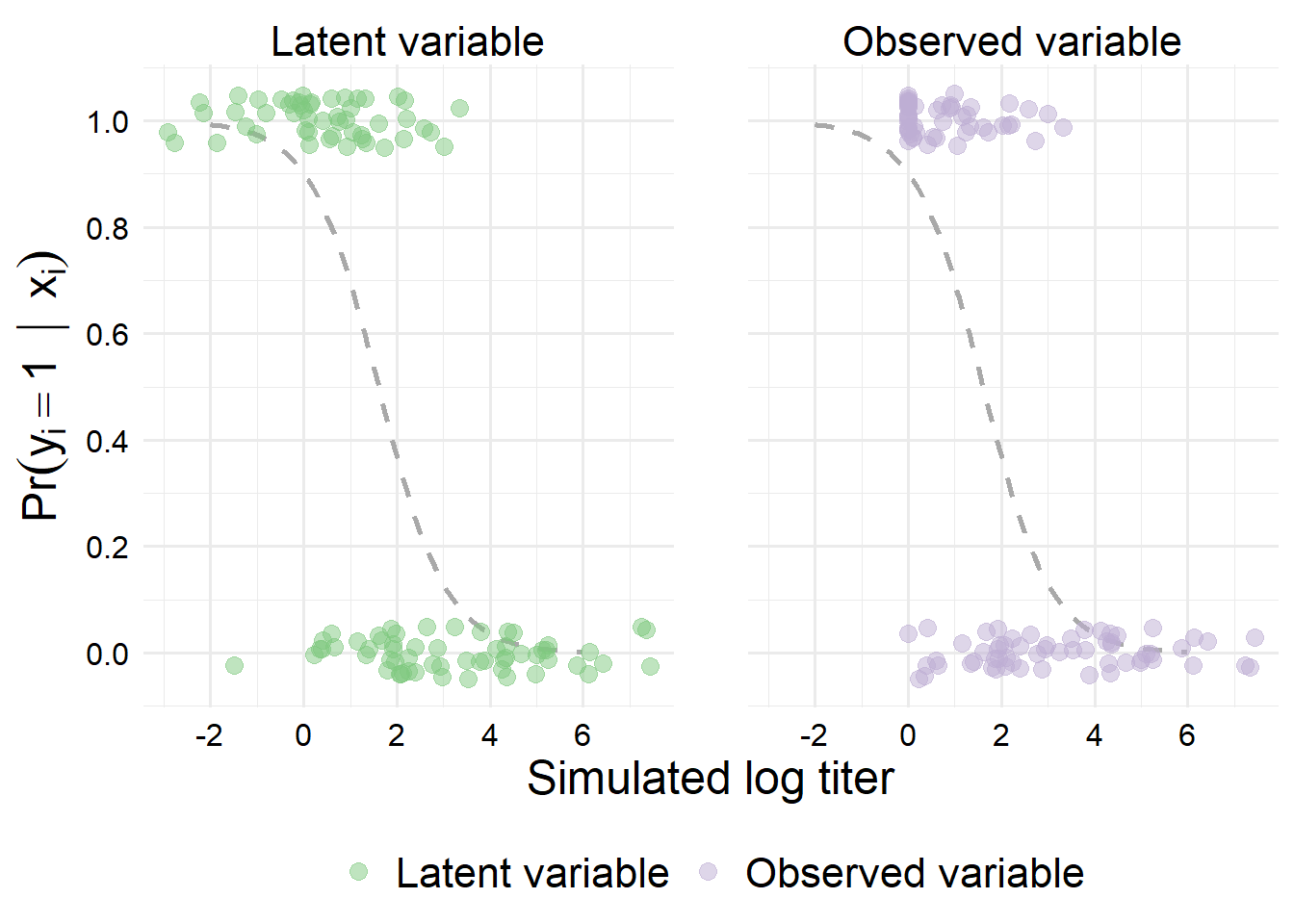
data_list <-
sim_data |>
dplyr::mutate(x_l = sim_parms$LoD) |>
dplyr::select(x, x_l, y) |>
as.list()
data_list$N <- sim_parms$n
str(data_list)List of 4
$ x : num [1:110] 0.123 1.872 0.932 1.865 1.925 ...
$ x_l: num [1:110] 0 0 0 0 0 0 0 0 0 0 ...
$ y : int [1:110] 1 0 1 0 0 0 1 0 0 0 ...
$ N : num 110mod_pth <- here::here(pth_base, "Ex4a.stan")
mod <- cmdstanr::cmdstan_model(mod_pth, compile = FALSE)
mod$compile(force_recompile = TRUE, pedantic = TRUE)In file included from stan/lib/stan_math/stan/math/prim/prob/von_mises_lccdf.hpp:5,
from stan/lib/stan_math/stan/math/prim/prob/von_mises_ccdf_log.hpp:4,
from stan/lib/stan_math/stan/math/prim/prob.hpp:356,
from stan/lib/stan_math/stan/math/prim.hpp:16,
from stan/lib/stan_math/stan/math/rev.hpp:14,
from stan/lib/stan_math/stan/math.hpp:19,
from stan/src/stan/model/model_header.hpp:4,
from C:/Users/Zane/AppData/Local/Temp/Rtmp0aU2FX/model-374c30036286.hpp:2:
stan/lib/stan_math/stan/math/prim/prob/von_mises_cdf.hpp: In function 'stan::return_type_t<T_x, T_sigma, T_l> stan::math::von_mises_cdf(const T_x&, const T_mu&, const T_k&)':
stan/lib/stan_math/stan/math/prim/prob/von_mises_cdf.hpp:194: note: '-Wmisleading-indentation' is disabled from this point onwards, since column-tracking was disabled due to the size of the code/headers
194 | if (cdf_n < 0.0)
| stan/lib/stan_math/stan/math/prim/prob/von_mises_cdf.hpp:194: note: adding '-flarge-source-files' will allow for more column-tracking support, at the expense of compilation time and memoryfit <- mod$sample(
data_list,
seed = 25452345,
parallel_chains = 4,
iter_warmup = 500,
iter_sampling = 2500,
show_messages = T
)Running MCMC with 4 parallel chains...
Chain 1 Iteration: 1 / 3000 [ 0%] (Warmup)
Chain 1 Iteration: 100 / 3000 [ 3%] (Warmup)
Chain 1 Iteration: 200 / 3000 [ 6%] (Warmup)
Chain 1 Iteration: 300 / 3000 [ 10%] (Warmup)
Chain 1 Iteration: 400 / 3000 [ 13%] (Warmup)
Chain 1 Iteration: 500 / 3000 [ 16%] (Warmup)
Chain 1 Iteration: 501 / 3000 [ 16%] (Sampling)
Chain 1 Iteration: 600 / 3000 [ 20%] (Sampling)
Chain 1 Iteration: 700 / 3000 [ 23%] (Sampling)
Chain 1 Iteration: 800 / 3000 [ 26%] (Sampling)
Chain 1 Iteration: 900 / 3000 [ 30%] (Sampling)
Chain 1 Iteration: 1000 / 3000 [ 33%] (Sampling)
Chain 2 Iteration: 1 / 3000 [ 0%] (Warmup)
Chain 2 Iteration: 100 / 3000 [ 3%] (Warmup)
Chain 2 Iteration: 200 / 3000 [ 6%] (Warmup)
Chain 2 Iteration: 300 / 3000 [ 10%] (Warmup)
Chain 2 Iteration: 400 / 3000 [ 13%] (Warmup)
Chain 2 Iteration: 500 / 3000 [ 16%] (Warmup)
Chain 2 Iteration: 501 / 3000 [ 16%] (Sampling)
Chain 2 Iteration: 600 / 3000 [ 20%] (Sampling)
Chain 2 Iteration: 700 / 3000 [ 23%] (Sampling)
Chain 2 Iteration: 800 / 3000 [ 26%] (Sampling)
Chain 2 Iteration: 900 / 3000 [ 30%] (Sampling)
Chain 3 Iteration: 1 / 3000 [ 0%] (Warmup)
Chain 3 Iteration: 100 / 3000 [ 3%] (Warmup)
Chain 3 Iteration: 200 / 3000 [ 6%] (Warmup)
Chain 3 Iteration: 300 / 3000 [ 10%] (Warmup)
Chain 3 Iteration: 400 / 3000 [ 13%] (Warmup)
Chain 3 Iteration: 500 / 3000 [ 16%] (Warmup)
Chain 3 Iteration: 501 / 3000 [ 16%] (Sampling)
Chain 3 Iteration: 600 / 3000 [ 20%] (Sampling)
Chain 3 Iteration: 700 / 3000 [ 23%] (Sampling)
Chain 3 Iteration: 800 / 3000 [ 26%] (Sampling)
Chain 4 Iteration: 1 / 3000 [ 0%] (Warmup)
Chain 4 Iteration: 100 / 3000 [ 3%] (Warmup)
Chain 4 Iteration: 200 / 3000 [ 6%] (Warmup)
Chain 4 Iteration: 300 / 3000 [ 10%] (Warmup)
Chain 4 Iteration: 400 / 3000 [ 13%] (Warmup)
Chain 4 Iteration: 500 / 3000 [ 16%] (Warmup)
Chain 4 Iteration: 501 / 3000 [ 16%] (Sampling)
Chain 4 Iteration: 600 / 3000 [ 20%] (Sampling) Chain 4 Informational Message: The current Metropolis proposal is about to be rejected because of the following issue:Chain 4 Exception: normal_lpdf: Scale parameter is 0, but must be positive! (in 'C:/Users/Zane/AppData/Local/Temp/Rtmp0aU2FX/model-374c30036286.stan', line 59, column 3 to column 32)Chain 4 If this warning occurs sporadically, such as for highly constrained variable types like covariance matrices, then the sampler is fine,Chain 4 but if this warning occurs often then your model may be either severely ill-conditioned or misspecified.Chain 4 Chain 1 Iteration: 1100 / 3000 [ 36%] (Sampling)
Chain 1 Iteration: 1200 / 3000 [ 40%] (Sampling)
Chain 1 Iteration: 1300 / 3000 [ 43%] (Sampling)
Chain 1 Iteration: 1400 / 3000 [ 46%] (Sampling)
Chain 1 Iteration: 1500 / 3000 [ 50%] (Sampling)
Chain 1 Iteration: 1600 / 3000 [ 53%] (Sampling)
Chain 1 Iteration: 1700 / 3000 [ 56%] (Sampling)
Chain 1 Iteration: 1800 / 3000 [ 60%] (Sampling)
Chain 1 Iteration: 1900 / 3000 [ 63%] (Sampling)
Chain 1 Iteration: 2000 / 3000 [ 66%] (Sampling)
Chain 1 Iteration: 2100 / 3000 [ 70%] (Sampling)
Chain 1 Iteration: 2200 / 3000 [ 73%] (Sampling)
Chain 1 Iteration: 2300 / 3000 [ 76%] (Sampling)
Chain 1 Iteration: 2400 / 3000 [ 80%] (Sampling)
Chain 1 Iteration: 2500 / 3000 [ 83%] (Sampling)
Chain 1 Iteration: 2600 / 3000 [ 86%] (Sampling)
Chain 1 Iteration: 2700 / 3000 [ 90%] (Sampling)
Chain 1 Iteration: 2800 / 3000 [ 93%] (Sampling)
Chain 1 Iteration: 2900 / 3000 [ 96%] (Sampling)
Chain 1 Iteration: 3000 / 3000 [100%] (Sampling)
Chain 1 finished in 0.4 seconds.
Chain 2 Iteration: 1000 / 3000 [ 33%] (Sampling)
Chain 2 Iteration: 1100 / 3000 [ 36%] (Sampling)
Chain 2 Iteration: 1200 / 3000 [ 40%] (Sampling)
Chain 2 Iteration: 1300 / 3000 [ 43%] (Sampling)
Chain 2 Iteration: 1400 / 3000 [ 46%] (Sampling)
Chain 2 Iteration: 1500 / 3000 [ 50%] (Sampling)
Chain 2 Iteration: 1600 / 3000 [ 53%] (Sampling)
Chain 2 Iteration: 1700 / 3000 [ 56%] (Sampling)
Chain 2 Iteration: 1800 / 3000 [ 60%] (Sampling)
Chain 2 Iteration: 1900 / 3000 [ 63%] (Sampling)
Chain 2 Iteration: 2000 / 3000 [ 66%] (Sampling)
Chain 2 Iteration: 2100 / 3000 [ 70%] (Sampling)
Chain 2 Iteration: 2200 / 3000 [ 73%] (Sampling)
Chain 2 Iteration: 2300 / 3000 [ 76%] (Sampling)
Chain 2 Iteration: 2400 / 3000 [ 80%] (Sampling)
Chain 2 Iteration: 2500 / 3000 [ 83%] (Sampling)
Chain 2 Iteration: 2600 / 3000 [ 86%] (Sampling)
Chain 2 Iteration: 2700 / 3000 [ 90%] (Sampling)
Chain 2 Iteration: 2800 / 3000 [ 93%] (Sampling)
Chain 2 Iteration: 2900 / 3000 [ 96%] (Sampling)
Chain 2 Iteration: 3000 / 3000 [100%] (Sampling)
Chain 2 finished in 0.3 seconds.
Chain 3 Iteration: 900 / 3000 [ 30%] (Sampling)
Chain 3 Iteration: 1000 / 3000 [ 33%] (Sampling)
Chain 3 Iteration: 1100 / 3000 [ 36%] (Sampling)
Chain 3 Iteration: 1200 / 3000 [ 40%] (Sampling)
Chain 3 Iteration: 1300 / 3000 [ 43%] (Sampling)
Chain 3 Iteration: 1400 / 3000 [ 46%] (Sampling)
Chain 3 Iteration: 1500 / 3000 [ 50%] (Sampling)
Chain 3 Iteration: 1600 / 3000 [ 53%] (Sampling)
Chain 3 Iteration: 1700 / 3000 [ 56%] (Sampling)
Chain 3 Iteration: 1800 / 3000 [ 60%] (Sampling)
Chain 3 Iteration: 1900 / 3000 [ 63%] (Sampling)
Chain 3 Iteration: 2000 / 3000 [ 66%] (Sampling)
Chain 3 Iteration: 2100 / 3000 [ 70%] (Sampling)
Chain 3 Iteration: 2200 / 3000 [ 73%] (Sampling)
Chain 3 Iteration: 2300 / 3000 [ 76%] (Sampling)
Chain 3 Iteration: 2400 / 3000 [ 80%] (Sampling)
Chain 3 Iteration: 2500 / 3000 [ 83%] (Sampling)
Chain 3 Iteration: 2600 / 3000 [ 86%] (Sampling)
Chain 3 Iteration: 2700 / 3000 [ 90%] (Sampling)
Chain 3 Iteration: 2800 / 3000 [ 93%] (Sampling)
Chain 3 Iteration: 2900 / 3000 [ 96%] (Sampling)
Chain 3 Iteration: 3000 / 3000 [100%] (Sampling)
Chain 3 finished in 0.4 seconds.
Chain 4 Iteration: 700 / 3000 [ 23%] (Sampling)
Chain 4 Iteration: 800 / 3000 [ 26%] (Sampling)
Chain 4 Iteration: 900 / 3000 [ 30%] (Sampling)
Chain 4 Iteration: 1000 / 3000 [ 33%] (Sampling)
Chain 4 Iteration: 1100 / 3000 [ 36%] (Sampling)
Chain 4 Iteration: 1200 / 3000 [ 40%] (Sampling)
Chain 4 Iteration: 1300 / 3000 [ 43%] (Sampling)
Chain 4 Iteration: 1400 / 3000 [ 46%] (Sampling)
Chain 4 Iteration: 1500 / 3000 [ 50%] (Sampling)
Chain 4 Iteration: 1600 / 3000 [ 53%] (Sampling)
Chain 4 Iteration: 1700 / 3000 [ 56%] (Sampling)
Chain 4 Iteration: 1800 / 3000 [ 60%] (Sampling)
Chain 4 Iteration: 1900 / 3000 [ 63%] (Sampling)
Chain 4 Iteration: 2000 / 3000 [ 66%] (Sampling)
Chain 4 Iteration: 2100 / 3000 [ 70%] (Sampling)
Chain 4 Iteration: 2200 / 3000 [ 73%] (Sampling)
Chain 4 Iteration: 2300 / 3000 [ 76%] (Sampling)
Chain 4 Iteration: 2400 / 3000 [ 80%] (Sampling)
Chain 4 Iteration: 2500 / 3000 [ 83%] (Sampling)
Chain 4 Iteration: 2600 / 3000 [ 86%] (Sampling)
Chain 4 Iteration: 2700 / 3000 [ 90%] (Sampling)
Chain 4 Iteration: 2800 / 3000 [ 93%] (Sampling)
Chain 4 Iteration: 2900 / 3000 [ 96%] (Sampling)
Chain 4 Iteration: 3000 / 3000 [100%] (Sampling)
Chain 4 finished in 0.4 seconds.
All 4 chains finished successfully.
Mean chain execution time: 0.4 seconds.
Total execution time: 0.6 seconds.fit$cmdstan_diagnose()Processing csv files: C:/Users/Zane/AppData/Local/Temp/Rtmp0aU2FX/Ex4a-202403032225-1-7db877.csv, C:/Users/Zane/AppData/Local/Temp/Rtmp0aU2FX/Ex4a-202403032225-2-7db877.csv, C:/Users/Zane/AppData/Local/Temp/Rtmp0aU2FX/Ex4a-202403032225-3-7db877.csv, C:/Users/Zane/AppData/Local/Temp/Rtmp0aU2FX/Ex4a-202403032225-4-7db877.csv
Checking sampler transitions treedepth.
Treedepth satisfactory for all transitions.
Checking sampler transitions for divergences.
No divergent transitions found.
Checking E-BFMI - sampler transitions HMC potential energy.
E-BFMI satisfactory.
Effective sample size satisfactory.
Split R-hat values satisfactory all parameters.
Processing complete, no problems detected.fit$summary()# A tibble: 5 × 10
variable mean median sd mad q5 q95 rhat ess_bulk ess_tail
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 lp__ -190. -189. 1.44 1.21 -192. -188. 1.00 4408. 5589.
2 alpha 1.91 1.90 0.411 0.416 1.26 2.60 1.00 5508. 5501.
3 beta -1.23 -1.22 0.226 0.226 -1.62 -0.881 1.00 5323. 5512.
4 mu_x 1.84 1.85 0.234 0.231 1.45 2.22 1.00 8886. 6064.
5 sigma_x 2.32 2.31 0.177 0.174 2.05 2.63 1.00 9161. 6999.6.2 Effect of vaccine
Of course, a more interesting question is when we have
In this framework,
Given the generative model, we can simulate data which follow our assumptions.
set.seed(341341)
# Some parameters are commented out because I originally had a global
# intercept for mu and for p, but then the intercept parameters are
# nonidentifiable under index coding as written.
sim2_parms <- list(
n = 116,
#a0 = 2,
a1 = c(2.5, 4),
#b0 = 1.5,
b1 = c(1.7, 2.2),
b2 = c(-0.67, -1.37),
sigma_x = 1.5,
LoD = 3,
latent = TRUE
)
sim_two_groups <- function(n, b1, b2, a1, sigma_x, LoD,
latent = TRUE) {
out <- tibble::tibble(
# Randomly assign each individual to 1 (placebo) or 2 (vaccine)
t = rbinom(n, size = 1, prob = 0.5) + 1,
mu = a1[t],
x_star = rnorm(n, mu, sigma_x),
x = dplyr::if_else(x_star < LoD, 0.5 * LoD, x_star),
p = inv_logit(b1[t] + b2[t] * x_star),
y = rbinom(n, 1, prob = p)
)
# If the arg 'latent' is specified as anything other than FALSE, return the
# latent variables that we don't observe. Otherwise return only (X, y).
if (isFALSE(latent)) {
out <- out |> dplyr::select(t, x, y)
}
return(out)
}
sim_data_4b <- do.call(sim_two_groups, sim2_parms)tab_dat <-
sim_data_4b |>
dplyr::mutate(
t = factor(
t,
levels = c(2, 1),
labels = c("Vaccine", "Placebo")
),
y = factor(
y,
levels = c(1, 0),
labels = c("Infected", "Not infected")
)
)
tab_dat |>
gtsummary::tbl_cross(
row = t, col = y,
label = list(t ~ "Treatment", y ~ "Outcome")
)| Outcome | Total | ||
|---|---|---|---|
| Infected | Not infected | ||
| Treatment | |||
| Vaccine | 5 | 49 | 54 |
| Placebo | 28 | 34 | 62 |
| Total | 33 | 83 | 116 |
Because the data are from a (hypothetical) clinical trial, the typical epidemiological approach to data analysis, if we do not care about the effect of the mediator
tab <- table(tab_dat$t, tab_dat$y, dnn = c("Treatment", "Outcome"))
epiR_out <- epiR::epi.2by2(
tab,
method = "cohort.count"
)
epiR_out Outcome + Outcome - Total Inc risk *
Exposed + 5 49 54 9.26 (3.08 to 20.30)
Exposed - 28 34 62 45.16 (32.48 to 58.32)
Total 33 83 116 28.45 (20.46 to 37.57)
Point estimates and 95% CIs:
-------------------------------------------------------------------
Inc risk ratio 0.21 (0.09, 0.49)
Inc odds ratio 0.12 (0.04, 0.35)
Attrib risk in the exposed * -35.90 (-50.50, -21.30)
Attrib fraction in the exposed (%) -387.74 (-1074.55, -102.54)
Attrib risk in the population * -16.71 (-31.57, -1.85)
Attrib fraction in the population (%) -58.75 (-89.23, -33.18)
-------------------------------------------------------------------
Uncorrected chi2 test that OR = 1: chi2(1) = 18.276 Pr>chi2 = <0.001
Fisher exact test that OR = 1: Pr>chi2 = <0.001
Wald confidence limits
CI: confidence interval
* Outcomes per 100 population units # This one takes the variables in the opposite direction so easier to do it
# this way
epiDisplay::csi(
caseexp = tab[[1]],
controlex = tab[[3]],
casenonex = tab[[2]],
controlnonex = tab[[4]]
)
Exposure
Outcome Non-exposed Exposed Total
Negative 34 49 83
Positive 28 5 33
Total 62 54 116
Rne Re Rt
Risk 0.45 0.09 0.28
Estimate Lower95ci Upper95ci
Risk difference (Re - Rne) -0.36 -0.5 -0.2
Risk ratio 0.21 0.1 0.45
Protective efficacy =(Rne-Re)/Rne*100 79.5 55.46 90.1
or percent of risk reduced
Number needed to treat (NNT) 2.79 2 4.99
or -1/(risk difference) So if we didn’t care about the effect of $ and a 95% CI of $\left(r round(epiR_out
However, in our study we specifically want to know how much of the lower risk is explained by the antibody titer, and how much is not. This analysis is more complicated, and requires us to use a regression model. Fortunately we know the data generating process, so writing the Stan code for an accurate model is not too hard.
mod_pth <- here::here(pth_base, "Ex4b.stan")
mod4b <- cmdstanr::cmdstan_model(mod_pth, compile = F)
mod4b$compile(pedantic = TRUE, force_recompile = TRUE)In file included from stan/lib/stan_math/stan/math/prim/prob/von_mises_lccdf.hpp:5,
from stan/lib/stan_math/stan/math/prim/prob/von_mises_ccdf_log.hpp:4,
from stan/lib/stan_math/stan/math/prim/prob.hpp:356,
from stan/lib/stan_math/stan/math/prim.hpp:16,
from stan/lib/stan_math/stan/math/rev.hpp:14,
from stan/lib/stan_math/stan/math.hpp:19,
from stan/src/stan/model/model_header.hpp:4,
from C:/Users/Zane/AppData/Local/Temp/Rtmp0aU2FX/model-374c53c17b4c.hpp:2:
stan/lib/stan_math/stan/math/prim/prob/von_mises_cdf.hpp: In function 'stan::return_type_t<T_x, T_sigma, T_l> stan::math::von_mises_cdf(const T_x&, const T_mu&, const T_k&)':
stan/lib/stan_math/stan/math/prim/prob/von_mises_cdf.hpp:194: note: '-Wmisleading-indentation' is disabled from this point onwards, since column-tracking was disabled due to the size of the code/headers
194 | if (cdf_n < 0.0)
| stan/lib/stan_math/stan/math/prim/prob/von_mises_cdf.hpp:194: note: adding '-flarge-source-files' will allow for more column-tracking support, at the expense of compilation time and memorymod4b_data <- sim_data_4b |>
dplyr::mutate(x_l = sim2_parms$LoD, t = as.integer(t)) |>
dplyr::select(t, y, x, x_l) |>
as.list()
mod4b_data <- c(
"N" = nrow(sim_data_4b),
"k" = as.integer(max(mod4b_data$t)),
mod4b_data
)
str(mod4b_data)List of 6
$ N : int 116
$ k : int 2
$ t : int [1:116] 1 1 2 1 1 1 2 2 2 2 ...
$ y : int [1:116] 1 0 0 0 0 0 0 0 0 0 ...
$ x : num [1:116] 1.5 3.19 1.5 3.79 4.7 ...
$ x_l: num [1:116] 3 3 3 3 3 3 3 3 3 3 ...paste0(
"Naruto checked the data and he says:\n",
round(mean(mod4b_data$x <= mod4b_data$x_l), 4) * 100,
"% of x values are below the LoD!\nBelieve it!"
) |> cat()Naruto checked the data and he says:
46.55% of x values are below the LoD!
Believe it!fit4b <- mod4b$sample(
mod4b_data,
seed = 5234521,
parallel_chains = 4,
iter_warmup = 500,
iter_sampling = 2500,
show_messages = T
)Running MCMC with 4 parallel chains...
Chain 1 Iteration: 1 / 3000 [ 0%] (Warmup)
Chain 1 Iteration: 100 / 3000 [ 3%] (Warmup) Chain 1 Informational Message: The current Metropolis proposal is about to be rejected because of the following issue:Chain 1 Exception: normal_lcdf: Scale parameter is 0, but must be positive! (in 'C:/Users/Zane/AppData/Local/Temp/Rtmp0aU2FX/model-374c53c17b4c.stan', line 75, column 3 to column 50)Chain 1 If this warning occurs sporadically, such as for highly constrained variable types like covariance matrices, then the sampler is fine,Chain 1 but if this warning occurs often then your model may be either severely ill-conditioned or misspecified.Chain 1 Chain 2 Iteration: 1 / 3000 [ 0%] (Warmup)
Chain 2 Iteration: 100 / 3000 [ 3%] (Warmup)
Chain 3 Iteration: 1 / 3000 [ 0%] (Warmup)
Chain 3 Iteration: 100 / 3000 [ 3%] (Warmup)
Chain 4 Iteration: 1 / 3000 [ 0%] (Warmup)
Chain 4 Iteration: 100 / 3000 [ 3%] (Warmup) Chain 4 Informational Message: The current Metropolis proposal is about to be rejected because of the following issue:Chain 4 Exception: normal_lcdf: Scale parameter is 0, but must be positive! (in 'C:/Users/Zane/AppData/Local/Temp/Rtmp0aU2FX/model-374c53c17b4c.stan', line 75, column 3 to column 50)Chain 4 If this warning occurs sporadically, such as for highly constrained variable types like covariance matrices, then the sampler is fine,Chain 4 but if this warning occurs often then your model may be either severely ill-conditioned or misspecified.Chain 4 Chain 1 Iteration: 200 / 3000 [ 6%] (Warmup)
Chain 1 Iteration: 300 / 3000 [ 10%] (Warmup)
Chain 1 Iteration: 400 / 3000 [ 13%] (Warmup)
Chain 1 Iteration: 500 / 3000 [ 16%] (Warmup)
Chain 1 Iteration: 501 / 3000 [ 16%] (Sampling)
Chain 2 Iteration: 200 / 3000 [ 6%] (Warmup)
Chain 2 Iteration: 300 / 3000 [ 10%] (Warmup)
Chain 2 Iteration: 400 / 3000 [ 13%] (Warmup)
Chain 2 Iteration: 500 / 3000 [ 16%] (Warmup)
Chain 2 Iteration: 501 / 3000 [ 16%] (Sampling)
Chain 3 Iteration: 200 / 3000 [ 6%] (Warmup)
Chain 3 Iteration: 300 / 3000 [ 10%] (Warmup)
Chain 3 Iteration: 400 / 3000 [ 13%] (Warmup)
Chain 3 Iteration: 500 / 3000 [ 16%] (Warmup)
Chain 3 Iteration: 501 / 3000 [ 16%] (Sampling)
Chain 4 Iteration: 200 / 3000 [ 6%] (Warmup)
Chain 4 Iteration: 300 / 3000 [ 10%] (Warmup)
Chain 4 Iteration: 400 / 3000 [ 13%] (Warmup)
Chain 1 Iteration: 600 / 3000 [ 20%] (Sampling)
Chain 1 Iteration: 700 / 3000 [ 23%] (Sampling)
Chain 2 Iteration: 600 / 3000 [ 20%] (Sampling)
Chain 2 Iteration: 700 / 3000 [ 23%] (Sampling)
Chain 3 Iteration: 600 / 3000 [ 20%] (Sampling)
Chain 3 Iteration: 700 / 3000 [ 23%] (Sampling)
Chain 4 Iteration: 500 / 3000 [ 16%] (Warmup)
Chain 4 Iteration: 501 / 3000 [ 16%] (Sampling)
Chain 4 Iteration: 600 / 3000 [ 20%] (Sampling)
Chain 4 Iteration: 700 / 3000 [ 23%] (Sampling)
Chain 1 Iteration: 800 / 3000 [ 26%] (Sampling)
Chain 1 Iteration: 900 / 3000 [ 30%] (Sampling)
Chain 1 Iteration: 1000 / 3000 [ 33%] (Sampling)
Chain 2 Iteration: 800 / 3000 [ 26%] (Sampling)
Chain 2 Iteration: 900 / 3000 [ 30%] (Sampling)
Chain 2 Iteration: 1000 / 3000 [ 33%] (Sampling)
Chain 3 Iteration: 800 / 3000 [ 26%] (Sampling)
Chain 3 Iteration: 900 / 3000 [ 30%] (Sampling)
Chain 3 Iteration: 1000 / 3000 [ 33%] (Sampling)
Chain 4 Iteration: 800 / 3000 [ 26%] (Sampling)
Chain 4 Iteration: 900 / 3000 [ 30%] (Sampling)
Chain 1 Iteration: 1100 / 3000 [ 36%] (Sampling)
Chain 1 Iteration: 1200 / 3000 [ 40%] (Sampling)
Chain 2 Iteration: 1100 / 3000 [ 36%] (Sampling)
Chain 3 Iteration: 1100 / 3000 [ 36%] (Sampling)
Chain 3 Iteration: 1200 / 3000 [ 40%] (Sampling)
Chain 4 Iteration: 1000 / 3000 [ 33%] (Sampling)
Chain 4 Iteration: 1100 / 3000 [ 36%] (Sampling)
Chain 1 Iteration: 1300 / 3000 [ 43%] (Sampling)
Chain 1 Iteration: 1400 / 3000 [ 46%] (Sampling)
Chain 2 Iteration: 1200 / 3000 [ 40%] (Sampling)
Chain 2 Iteration: 1300 / 3000 [ 43%] (Sampling)
Chain 2 Iteration: 1400 / 3000 [ 46%] (Sampling)
Chain 3 Iteration: 1300 / 3000 [ 43%] (Sampling)
Chain 3 Iteration: 1400 / 3000 [ 46%] (Sampling)
Chain 4 Iteration: 1200 / 3000 [ 40%] (Sampling)
Chain 4 Iteration: 1300 / 3000 [ 43%] (Sampling)
Chain 1 Iteration: 1500 / 3000 [ 50%] (Sampling)
Chain 1 Iteration: 1600 / 3000 [ 53%] (Sampling)
Chain 1 Iteration: 1700 / 3000 [ 56%] (Sampling)
Chain 2 Iteration: 1500 / 3000 [ 50%] (Sampling)
Chain 2 Iteration: 1600 / 3000 [ 53%] (Sampling)
Chain 3 Iteration: 1500 / 3000 [ 50%] (Sampling)
Chain 3 Iteration: 1600 / 3000 [ 53%] (Sampling)
Chain 3 Iteration: 1700 / 3000 [ 56%] (Sampling)
Chain 4 Iteration: 1400 / 3000 [ 46%] (Sampling)
Chain 4 Iteration: 1500 / 3000 [ 50%] (Sampling)
Chain 1 Iteration: 1800 / 3000 [ 60%] (Sampling)
Chain 1 Iteration: 1900 / 3000 [ 63%] (Sampling)
Chain 2 Iteration: 1700 / 3000 [ 56%] (Sampling)
Chain 2 Iteration: 1800 / 3000 [ 60%] (Sampling)
Chain 3 Iteration: 1800 / 3000 [ 60%] (Sampling)
Chain 3 Iteration: 1900 / 3000 [ 63%] (Sampling)
Chain 4 Iteration: 1600 / 3000 [ 53%] (Sampling)
Chain 4 Iteration: 1700 / 3000 [ 56%] (Sampling)
Chain 1 Iteration: 2000 / 3000 [ 66%] (Sampling)
Chain 1 Iteration: 2100 / 3000 [ 70%] (Sampling)
Chain 2 Iteration: 1900 / 3000 [ 63%] (Sampling)
Chain 3 Iteration: 2000 / 3000 [ 66%] (Sampling)
Chain 3 Iteration: 2100 / 3000 [ 70%] (Sampling)
Chain 4 Iteration: 1800 / 3000 [ 60%] (Sampling)
Chain 4 Iteration: 1900 / 3000 [ 63%] (Sampling)
Chain 1 Iteration: 2200 / 3000 [ 73%] (Sampling)
Chain 1 Iteration: 2300 / 3000 [ 76%] (Sampling)
Chain 2 Iteration: 2000 / 3000 [ 66%] (Sampling)
Chain 2 Iteration: 2100 / 3000 [ 70%] (Sampling)
Chain 2 Iteration: 2200 / 3000 [ 73%] (Sampling)
Chain 3 Iteration: 2200 / 3000 [ 73%] (Sampling)
Chain 3 Iteration: 2300 / 3000 [ 76%] (Sampling)
Chain 4 Iteration: 2000 / 3000 [ 66%] (Sampling)
Chain 4 Iteration: 2100 / 3000 [ 70%] (Sampling)
Chain 1 Iteration: 2400 / 3000 [ 80%] (Sampling)
Chain 2 Iteration: 2300 / 3000 [ 76%] (Sampling)
Chain 3 Iteration: 2400 / 3000 [ 80%] (Sampling)
Chain 3 Iteration: 2500 / 3000 [ 83%] (Sampling)
Chain 4 Iteration: 2200 / 3000 [ 73%] (Sampling)
Chain 1 Iteration: 2500 / 3000 [ 83%] (Sampling)
Chain 1 Iteration: 2600 / 3000 [ 86%] (Sampling)
Chain 2 Iteration: 2400 / 3000 [ 80%] (Sampling)
Chain 2 Iteration: 2500 / 3000 [ 83%] (Sampling)
Chain 3 Iteration: 2600 / 3000 [ 86%] (Sampling)
Chain 3 Iteration: 2700 / 3000 [ 90%] (Sampling)
Chain 3 Iteration: 2800 / 3000 [ 93%] (Sampling)
Chain 4 Iteration: 2300 / 3000 [ 76%] (Sampling)
Chain 4 Iteration: 2400 / 3000 [ 80%] (Sampling)
Chain 4 Iteration: 2500 / 3000 [ 83%] (Sampling)
Chain 1 Iteration: 2700 / 3000 [ 90%] (Sampling)
Chain 1 Iteration: 2800 / 3000 [ 93%] (Sampling)
Chain 1 Iteration: 2900 / 3000 [ 96%] (Sampling)
Chain 2 Iteration: 2600 / 3000 [ 86%] (Sampling)
Chain 2 Iteration: 2700 / 3000 [ 90%] (Sampling)
Chain 3 Iteration: 2900 / 3000 [ 96%] (Sampling)
Chain 4 Iteration: 2600 / 3000 [ 86%] (Sampling)
Chain 1 Iteration: 3000 / 3000 [100%] (Sampling)
Chain 2 Iteration: 2800 / 3000 [ 93%] (Sampling)
Chain 2 Iteration: 2900 / 3000 [ 96%] (Sampling)
Chain 3 Iteration: 3000 / 3000 [100%] (Sampling)
Chain 4 Iteration: 2700 / 3000 [ 90%] (Sampling)
Chain 4 Iteration: 2800 / 3000 [ 93%] (Sampling)
Chain 1 finished in 1.7 seconds.
Chain 3 finished in 1.6 seconds.
Chain 2 Iteration: 3000 / 3000 [100%] (Sampling)
Chain 4 Iteration: 2900 / 3000 [ 96%] (Sampling)
Chain 4 Iteration: 3000 / 3000 [100%] (Sampling)
Chain 2 finished in 1.8 seconds.
Chain 4 finished in 1.8 seconds.
All 4 chains finished successfully.
Mean chain execution time: 1.7 seconds.
Total execution time: 1.9 seconds.fit4b$summary()# A tibble: 8 × 10
variable mean median sd mad q5 q95 rhat ess_bulk
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 lp__ -157. -157. 1.95 1.85 -161. -154. 1.00 4215.
2 alpha_1[1] 2.45 2.46 0.292 0.284 1.95 2.90 1.00 8685.
3 alpha_1[2] 3.85 3.85 0.254 0.256 3.42 4.26 1.00 9349.
4 sigma_x 1.75 1.74 0.181 0.175 1.48 2.07 1.00 8116.
5 beta_1[1] 0.760 0.758 0.530 0.536 -0.102 1.65 1.00 6824.
6 beta_1[2] 1.45 1.40 1.17 1.15 -0.351 3.47 1.00 5524.
7 beta_2[1] -0.397 -0.392 0.197 0.197 -0.732 -0.0818 1.00 6880.
8 beta_2[2] -1.69 -1.61 0.666 0.640 -2.94 -0.752 1.00 5384.
# ℹ 1 more variable: ess_tail <dbl>str(sim2_parms)List of 7
$ n : num 116
$ a1 : num [1:2] 2.5 4
$ b1 : num [1:2] 1.7 2.2
$ b2 : num [1:2] -0.67 -1.37
$ sigma_x: num 1.5
$ LoD : num 3
$ latent : logi TRUEfit_summary <- fit4b$summary() |>
dplyr::select(variable, median, q5, q95) |>
dplyr::filter(variable != "lp__") |>
dplyr::mutate(
truth = c(
#sim2_parms$a0,
sim2_parms$a1[[1]],
sim2_parms$a1[[2]],
sim2_parms$sigma_x,
#sim2_parms$b0,
sim2_parms$b1[[1]],
sim2_parms$b1[[2]],
sim2_parms$b2[[1]],
sim2_parms$b2[[2]]
)
)
po <-
ggplot(fit_summary) +
aes(x = variable, y = median, ymin = q5, ymax = q95) +
geom_hline(yintercept = 0, linetype = "dashed", color = "gray") +
geom_pointrange() +
geom_point(aes(y = truth), shape = 4, color = "red", size = 3, stroke = 1) +
labs(
x = NULL,
y = "Parameter value",
title = "Model-estimated median with 95% CI; x marks true simulation value",
subtitle = "Estimated with observed (censored) values with correction"
)6.3 Model if x was not censored
mod4b_data_l <- sim_data_4b |>
dplyr::mutate(x_l = -9999, t = as.integer(t)) |>
dplyr::select(t, y, x = x_star, x_l) |>
as.list()
mod4b_data_l <- c(
"N" = nrow(sim_data_4b),
"k" = as.integer(max(mod4b_data_l$t)),
mod4b_data_l
)
str(mod4b_data_l)List of 6
$ N : int 116
$ k : int 2
$ t : int [1:116] 1 1 2 1 1 1 2 2 2 2 ...
$ y : int [1:116] 1 0 0 0 0 0 0 0 0 0 ...
$ x : num [1:116] 2.07 3.19 2.2 3.79 4.7 ...
$ x_l: num [1:116] -9999 -9999 -9999 -9999 -9999 ...fit4b_l <- mod4b$sample(
mod4b_data_l,
seed = 5234521,
parallel_chains = 4,
iter_warmup = 500,
iter_sampling = 2500,
show_messages = T
)Running MCMC with 4 parallel chains...
Chain 1 Iteration: 1 / 3000 [ 0%] (Warmup)
Chain 1 Iteration: 100 / 3000 [ 3%] (Warmup)
Chain 2 Iteration: 1 / 3000 [ 0%] (Warmup)
Chain 2 Iteration: 100 / 3000 [ 3%] (Warmup)
Chain 3 Iteration: 1 / 3000 [ 0%] (Warmup)
Chain 3 Iteration: 100 / 3000 [ 3%] (Warmup)
Chain 4 Iteration: 1 / 3000 [ 0%] (Warmup)
Chain 4 Iteration: 100 / 3000 [ 3%] (Warmup)
Chain 1 Iteration: 200 / 3000 [ 6%] (Warmup)
Chain 1 Iteration: 300 / 3000 [ 10%] (Warmup)
Chain 1 Iteration: 400 / 3000 [ 13%] (Warmup)
Chain 1 Iteration: 500 / 3000 [ 16%] (Warmup)
Chain 1 Iteration: 501 / 3000 [ 16%] (Sampling)
Chain 1 Iteration: 600 / 3000 [ 20%] (Sampling)
Chain 2 Iteration: 200 / 3000 [ 6%] (Warmup)
Chain 2 Iteration: 300 / 3000 [ 10%] (Warmup)
Chain 2 Iteration: 400 / 3000 [ 13%] (Warmup)
Chain 2 Iteration: 500 / 3000 [ 16%] (Warmup)
Chain 2 Iteration: 501 / 3000 [ 16%] (Sampling)
Chain 2 Iteration: 600 / 3000 [ 20%] (Sampling)
Chain 3 Iteration: 200 / 3000 [ 6%] (Warmup)
Chain 3 Iteration: 300 / 3000 [ 10%] (Warmup)
Chain 3 Iteration: 400 / 3000 [ 13%] (Warmup)
Chain 3 Iteration: 500 / 3000 [ 16%] (Warmup)
Chain 3 Iteration: 501 / 3000 [ 16%] (Sampling)
Chain 3 Iteration: 600 / 3000 [ 20%] (Sampling)
Chain 4 Iteration: 200 / 3000 [ 6%] (Warmup)
Chain 4 Iteration: 300 / 3000 [ 10%] (Warmup)
Chain 4 Iteration: 400 / 3000 [ 13%] (Warmup)
Chain 4 Iteration: 500 / 3000 [ 16%] (Warmup)
Chain 4 Iteration: 501 / 3000 [ 16%] (Sampling)
Chain 1 Iteration: 700 / 3000 [ 23%] (Sampling)
Chain 1 Iteration: 800 / 3000 [ 26%] (Sampling)
Chain 1 Iteration: 900 / 3000 [ 30%] (Sampling)
Chain 2 Iteration: 700 / 3000 [ 23%] (Sampling)
Chain 2 Iteration: 800 / 3000 [ 26%] (Sampling)
Chain 2 Iteration: 900 / 3000 [ 30%] (Sampling)
Chain 3 Iteration: 700 / 3000 [ 23%] (Sampling)
Chain 3 Iteration: 800 / 3000 [ 26%] (Sampling)
Chain 3 Iteration: 900 / 3000 [ 30%] (Sampling)
Chain 4 Iteration: 600 / 3000 [ 20%] (Sampling)
Chain 4 Iteration: 700 / 3000 [ 23%] (Sampling)
Chain 4 Iteration: 800 / 3000 [ 26%] (Sampling)
Chain 1 Iteration: 1000 / 3000 [ 33%] (Sampling)
Chain 1 Iteration: 1100 / 3000 [ 36%] (Sampling)
Chain 2 Iteration: 1000 / 3000 [ 33%] (Sampling)
Chain 2 Iteration: 1100 / 3000 [ 36%] (Sampling)
Chain 3 Iteration: 1000 / 3000 [ 33%] (Sampling)
Chain 3 Iteration: 1100 / 3000 [ 36%] (Sampling)
Chain 4 Iteration: 900 / 3000 [ 30%] (Sampling)
Chain 4 Iteration: 1000 / 3000 [ 33%] (Sampling)
Chain 1 Iteration: 1200 / 3000 [ 40%] (Sampling)
Chain 1 Iteration: 1300 / 3000 [ 43%] (Sampling)
Chain 1 Iteration: 1400 / 3000 [ 46%] (Sampling)
Chain 2 Iteration: 1200 / 3000 [ 40%] (Sampling)
Chain 2 Iteration: 1300 / 3000 [ 43%] (Sampling)
Chain 3 Iteration: 1200 / 3000 [ 40%] (Sampling)
Chain 3 Iteration: 1300 / 3000 [ 43%] (Sampling)
Chain 4 Iteration: 1100 / 3000 [ 36%] (Sampling)
Chain 4 Iteration: 1200 / 3000 [ 40%] (Sampling)
Chain 1 Iteration: 1500 / 3000 [ 50%] (Sampling)
Chain 1 Iteration: 1600 / 3000 [ 53%] (Sampling)
Chain 2 Iteration: 1400 / 3000 [ 46%] (Sampling)
Chain 2 Iteration: 1500 / 3000 [ 50%] (Sampling)
Chain 3 Iteration: 1400 / 3000 [ 46%] (Sampling)
Chain 3 Iteration: 1500 / 3000 [ 50%] (Sampling)
Chain 4 Iteration: 1300 / 3000 [ 43%] (Sampling)
Chain 4 Iteration: 1400 / 3000 [ 46%] (Sampling)
Chain 1 Iteration: 1700 / 3000 [ 56%] (Sampling)
Chain 1 Iteration: 1800 / 3000 [ 60%] (Sampling)
Chain 1 Iteration: 1900 / 3000 [ 63%] (Sampling)
Chain 2 Iteration: 1600 / 3000 [ 53%] (Sampling)
Chain 2 Iteration: 1700 / 3000 [ 56%] (Sampling)
Chain 3 Iteration: 1600 / 3000 [ 53%] (Sampling)
Chain 3 Iteration: 1700 / 3000 [ 56%] (Sampling)
Chain 3 Iteration: 1800 / 3000 [ 60%] (Sampling)
Chain 4 Iteration: 1500 / 3000 [ 50%] (Sampling)
Chain 4 Iteration: 1600 / 3000 [ 53%] (Sampling)
Chain 1 Iteration: 2000 / 3000 [ 66%] (Sampling)
Chain 1 Iteration: 2100 / 3000 [ 70%] (Sampling)
Chain 2 Iteration: 1800 / 3000 [ 60%] (Sampling)
Chain 2 Iteration: 1900 / 3000 [ 63%] (Sampling)
Chain 3 Iteration: 1900 / 3000 [ 63%] (Sampling)
Chain 3 Iteration: 2000 / 3000 [ 66%] (Sampling)
Chain 4 Iteration: 1700 / 3000 [ 56%] (Sampling)
Chain 4 Iteration: 1800 / 3000 [ 60%] (Sampling)
Chain 1 Iteration: 2200 / 3000 [ 73%] (Sampling)
Chain 1 Iteration: 2300 / 3000 [ 76%] (Sampling)
Chain 2 Iteration: 2000 / 3000 [ 66%] (Sampling)
Chain 2 Iteration: 2100 / 3000 [ 70%] (Sampling)
Chain 2 Iteration: 2200 / 3000 [ 73%] (Sampling)
Chain 3 Iteration: 2100 / 3000 [ 70%] (Sampling)
Chain 3 Iteration: 2200 / 3000 [ 73%] (Sampling)
Chain 4 Iteration: 1900 / 3000 [ 63%] (Sampling)
Chain 4 Iteration: 2000 / 3000 [ 66%] (Sampling)
Chain 4 Iteration: 2100 / 3000 [ 70%] (Sampling)
Chain 1 Iteration: 2400 / 3000 [ 80%] (Sampling)
Chain 1 Iteration: 2500 / 3000 [ 83%] (Sampling)
Chain 1 Iteration: 2600 / 3000 [ 86%] (Sampling)
Chain 2 Iteration: 2300 / 3000 [ 76%] (Sampling)
Chain 2 Iteration: 2400 / 3000 [ 80%] (Sampling)
Chain 3 Iteration: 2300 / 3000 [ 76%] (Sampling)
Chain 3 Iteration: 2400 / 3000 [ 80%] (Sampling)
Chain 4 Iteration: 2200 / 3000 [ 73%] (Sampling)
Chain 4 Iteration: 2300 / 3000 [ 76%] (Sampling)
Chain 1 Iteration: 2700 / 3000 [ 90%] (Sampling)
Chain 1 Iteration: 2800 / 3000 [ 93%] (Sampling)
Chain 1 Iteration: 2900 / 3000 [ 96%] (Sampling)
Chain 2 Iteration: 2500 / 3000 [ 83%] (Sampling)
Chain 2 Iteration: 2600 / 3000 [ 86%] (Sampling)
Chain 2 Iteration: 2700 / 3000 [ 90%] (Sampling)
Chain 3 Iteration: 2500 / 3000 [ 83%] (Sampling)
Chain 3 Iteration: 2600 / 3000 [ 86%] (Sampling)
Chain 4 Iteration: 2400 / 3000 [ 80%] (Sampling)
Chain 4 Iteration: 2500 / 3000 [ 83%] (Sampling)
Chain 4 Iteration: 2600 / 3000 [ 86%] (Sampling)
Chain 1 Iteration: 3000 / 3000 [100%] (Sampling)
Chain 2 Iteration: 2800 / 3000 [ 93%] (Sampling)
Chain 2 Iteration: 2900 / 3000 [ 96%] (Sampling)
Chain 3 Iteration: 2700 / 3000 [ 90%] (Sampling)
Chain 3 Iteration: 2800 / 3000 [ 93%] (Sampling)
Chain 4 Iteration: 2700 / 3000 [ 90%] (Sampling)
Chain 4 Iteration: 2800 / 3000 [ 93%] (Sampling)
Chain 1 finished in 1.4 seconds.
Chain 2 Iteration: 3000 / 3000 [100%] (Sampling)
Chain 3 Iteration: 2900 / 3000 [ 96%] (Sampling)
Chain 3 Iteration: 3000 / 3000 [100%] (Sampling)
Chain 4 Iteration: 2900 / 3000 [ 96%] (Sampling)
Chain 4 Iteration: 3000 / 3000 [100%] (Sampling)
Chain 2 finished in 1.5 seconds.
Chain 3 finished in 1.5 seconds.
Chain 4 finished in 1.5 seconds.
All 4 chains finished successfully.
Mean chain execution time: 1.5 seconds.
Total execution time: 1.6 seconds.fit4b_l$summary()# A tibble: 8 × 10
variable mean median sd mad q5 q95 rhat ess_bulk
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 lp__ -166. -165. 1.87 1.73 -169. -163. 1.00 4661.
2 alpha_1[1] 2.63 2.64 0.204 0.203 2.30 2.97 1.00 11142.
3 alpha_1[2] 3.90 3.91 0.214 0.213 3.55 4.25 1.00 12564.
4 sigma_x 1.61 1.61 0.108 0.108 1.44 1.79 1.00 10927.
5 beta_1[1] 1.50 1.48 0.599 0.597 0.545 2.51 1.00 7405.
6 beta_1[2] 1.21 1.19 1.09 1.09 -0.530 3.06 1.00 7679.
7 beta_2[1] -0.673 -0.661 0.213 0.216 -1.04 -0.342 1.00 7502.
8 beta_2[2] -1.26 -1.22 0.449 0.446 -2.06 -0.603 1.00 7520.
# ℹ 1 more variable: ess_tail <dbl>str(sim2_parms)List of 7
$ n : num 116
$ a1 : num [1:2] 2.5 4
$ b1 : num [1:2] 1.7 2.2
$ b2 : num [1:2] -0.67 -1.37
$ sigma_x: num 1.5
$ LoD : num 3
$ latent : logi TRUEfit_summary_l <- fit4b_l$summary() |>
dplyr::select(variable, median, q5, q95) |>
dplyr::filter(variable != "lp__") |>
dplyr::mutate(
truth = c(
#sim2_parms$a0,
sim2_parms$a1[[1]],
sim2_parms$a1[[2]],
sim2_parms$sigma_x,
#sim2_parms$b0,
sim2_parms$b1[[1]],
sim2_parms$b1[[2]],
sim2_parms$b2[[1]],
sim2_parms$b2[[2]]
)
)
pl <-
ggplot(fit_summary_l) +
aes(x = variable, y = median, ymin = q5, ymax = q95) +
geom_hline(yintercept = 0, linetype = "dashed", color = "gray") +
geom_pointrange() +
geom_point(aes(y = truth), shape = 4, color = "red", size = 3, stroke = 1) +
labs(
x = NULL,
y = "Parameter value",
title = "Model-estimated median with 95% CI; x marks true simulation value",
subtitle = "Estimated using true latent values"
)6.4 Do it the naive way
mod4b_data_n <- sim_data_4b |>
dplyr::mutate(x_l = -9999, t = as.integer(t)) |>
dplyr::select(t, y, x = x, x_l) |>
as.list()
mod4b_data_n <- c(
"N" = nrow(sim_data_4b),
"k" = as.integer(max(mod4b_data_n$t)),
mod4b_data_n
)
str(mod4b_data_n)List of 6
$ N : int 116
$ k : int 2
$ t : int [1:116] 1 1 2 1 1 1 2 2 2 2 ...
$ y : int [1:116] 1 0 0 0 0 0 0 0 0 0 ...
$ x : num [1:116] 1.5 3.19 1.5 3.79 4.7 ...
$ x_l: num [1:116] -9999 -9999 -9999 -9999 -9999 ...fit4b_n <- mod4b$sample(
mod4b_data_n,
seed = 873215,
parallel_chains = 4,
iter_warmup = 500,
iter_sampling = 2500,
show_messages = T
)Running MCMC with 4 parallel chains...
Chain 1 Iteration: 1 / 3000 [ 0%] (Warmup)
Chain 1 Iteration: 100 / 3000 [ 3%] (Warmup)
Chain 2 Iteration: 1 / 3000 [ 0%] (Warmup)
Chain 2 Iteration: 100 / 3000 [ 3%] (Warmup)
Chain 3 Iteration: 1 / 3000 [ 0%] (Warmup)
Chain 3 Iteration: 100 / 3000 [ 3%] (Warmup)
Chain 4 Iteration: 1 / 3000 [ 0%] (Warmup)
Chain 4 Iteration: 100 / 3000 [ 3%] (Warmup)
Chain 1 Iteration: 200 / 3000 [ 6%] (Warmup)
Chain 1 Iteration: 300 / 3000 [ 10%] (Warmup)
Chain 1 Iteration: 400 / 3000 [ 13%] (Warmup)
Chain 1 Iteration: 500 / 3000 [ 16%] (Warmup)
Chain 1 Iteration: 501 / 3000 [ 16%] (Sampling)
Chain 1 Iteration: 600 / 3000 [ 20%] (Sampling)
Chain 2 Iteration: 200 / 3000 [ 6%] (Warmup)
Chain 2 Iteration: 300 / 3000 [ 10%] (Warmup)
Chain 2 Iteration: 400 / 3000 [ 13%] (Warmup)
Chain 2 Iteration: 500 / 3000 [ 16%] (Warmup)
Chain 2 Iteration: 501 / 3000 [ 16%] (Sampling)
Chain 2 Iteration: 600 / 3000 [ 20%] (Sampling)
Chain 3 Iteration: 200 / 3000 [ 6%] (Warmup)
Chain 3 Iteration: 300 / 3000 [ 10%] (Warmup)
Chain 3 Iteration: 400 / 3000 [ 13%] (Warmup)
Chain 3 Iteration: 500 / 3000 [ 16%] (Warmup)
Chain 3 Iteration: 501 / 3000 [ 16%] (Sampling)
Chain 3 Iteration: 600 / 3000 [ 20%] (Sampling)
Chain 4 Iteration: 200 / 3000 [ 6%] (Warmup)
Chain 4 Iteration: 300 / 3000 [ 10%] (Warmup)
Chain 4 Iteration: 400 / 3000 [ 13%] (Warmup)
Chain 4 Iteration: 500 / 3000 [ 16%] (Warmup)
Chain 4 Iteration: 501 / 3000 [ 16%] (Sampling)
Chain 1 Iteration: 700 / 3000 [ 23%] (Sampling)
Chain 1 Iteration: 800 / 3000 [ 26%] (Sampling)
Chain 1 Iteration: 900 / 3000 [ 30%] (Sampling)
Chain 2 Iteration: 700 / 3000 [ 23%] (Sampling)
Chain 2 Iteration: 800 / 3000 [ 26%] (Sampling)
Chain 2 Iteration: 900 / 3000 [ 30%] (Sampling)
Chain 3 Iteration: 700 / 3000 [ 23%] (Sampling)
Chain 3 Iteration: 800 / 3000 [ 26%] (Sampling)
Chain 3 Iteration: 900 / 3000 [ 30%] (Sampling)
Chain 4 Iteration: 600 / 3000 [ 20%] (Sampling)
Chain 4 Iteration: 700 / 3000 [ 23%] (Sampling)
Chain 4 Iteration: 800 / 3000 [ 26%] (Sampling)
Chain 1 Iteration: 1000 / 3000 [ 33%] (Sampling)
Chain 1 Iteration: 1100 / 3000 [ 36%] (Sampling)
Chain 2 Iteration: 1000 / 3000 [ 33%] (Sampling)
Chain 2 Iteration: 1100 / 3000 [ 36%] (Sampling)
Chain 3 Iteration: 1000 / 3000 [ 33%] (Sampling)
Chain 3 Iteration: 1100 / 3000 [ 36%] (Sampling)
Chain 4 Iteration: 900 / 3000 [ 30%] (Sampling)
Chain 4 Iteration: 1000 / 3000 [ 33%] (Sampling)
Chain 1 Iteration: 1200 / 3000 [ 40%] (Sampling)
Chain 1 Iteration: 1300 / 3000 [ 43%] (Sampling)
Chain 2 Iteration: 1200 / 3000 [ 40%] (Sampling)
Chain 2 Iteration: 1300 / 3000 [ 43%] (Sampling)
Chain 3 Iteration: 1200 / 3000 [ 40%] (Sampling)
Chain 3 Iteration: 1300 / 3000 [ 43%] (Sampling)
Chain 4 Iteration: 1100 / 3000 [ 36%] (Sampling)
Chain 4 Iteration: 1200 / 3000 [ 40%] (Sampling)
Chain 1 Iteration: 1400 / 3000 [ 46%] (Sampling)
Chain 1 Iteration: 1500 / 3000 [ 50%] (Sampling)
Chain 2 Iteration: 1400 / 3000 [ 46%] (Sampling)
Chain 2 Iteration: 1500 / 3000 [ 50%] (Sampling)
Chain 3 Iteration: 1400 / 3000 [ 46%] (Sampling)
Chain 3 Iteration: 1500 / 3000 [ 50%] (Sampling)
Chain 4 Iteration: 1300 / 3000 [ 43%] (Sampling)
Chain 4 Iteration: 1400 / 3000 [ 46%] (Sampling)
Chain 1 Iteration: 1600 / 3000 [ 53%] (Sampling)
Chain 1 Iteration: 1700 / 3000 [ 56%] (Sampling)
Chain 2 Iteration: 1600 / 3000 [ 53%] (Sampling)
Chain 2 Iteration: 1700 / 3000 [ 56%] (Sampling)
Chain 3 Iteration: 1600 / 3000 [ 53%] (Sampling)
Chain 3 Iteration: 1700 / 3000 [ 56%] (Sampling)
Chain 4 Iteration: 1500 / 3000 [ 50%] (Sampling)
Chain 4 Iteration: 1600 / 3000 [ 53%] (Sampling)
Chain 4 Iteration: 1700 / 3000 [ 56%] (Sampling)
Chain 1 Iteration: 1800 / 3000 [ 60%] (Sampling)
Chain 1 Iteration: 1900 / 3000 [ 63%] (Sampling)
Chain 2 Iteration: 1800 / 3000 [ 60%] (Sampling)
Chain 2 Iteration: 1900 / 3000 [ 63%] (Sampling)
Chain 2 Iteration: 2000 / 3000 [ 66%] (Sampling)
Chain 3 Iteration: 1800 / 3000 [ 60%] (Sampling)
Chain 3 Iteration: 1900 / 3000 [ 63%] (Sampling)
Chain 4 Iteration: 1800 / 3000 [ 60%] (Sampling)
Chain 4 Iteration: 1900 / 3000 [ 63%] (Sampling)
Chain 1 Iteration: 2000 / 3000 [ 66%] (Sampling)
Chain 1 Iteration: 2100 / 3000 [ 70%] (Sampling)
Chain 2 Iteration: 2100 / 3000 [ 70%] (Sampling)
Chain 2 Iteration: 2200 / 3000 [ 73%] (Sampling)
Chain 3 Iteration: 2000 / 3000 [ 66%] (Sampling)
Chain 3 Iteration: 2100 / 3000 [ 70%] (Sampling)
Chain 4 Iteration: 2000 / 3000 [ 66%] (Sampling)
Chain 4 Iteration: 2100 / 3000 [ 70%] (Sampling)
Chain 1 Iteration: 2200 / 3000 [ 73%] (Sampling)
Chain 1 Iteration: 2300 / 3000 [ 76%] (Sampling)
Chain 2 Iteration: 2300 / 3000 [ 76%] (Sampling)
Chain 2 Iteration: 2400 / 3000 [ 80%] (Sampling)
Chain 2 Iteration: 2500 / 3000 [ 83%] (Sampling)
Chain 3 Iteration: 2200 / 3000 [ 73%] (Sampling)
Chain 3 Iteration: 2300 / 3000 [ 76%] (Sampling)
Chain 4 Iteration: 2200 / 3000 [ 73%] (Sampling)
Chain 4 Iteration: 2300 / 3000 [ 76%] (Sampling)
Chain 1 Iteration: 2400 / 3000 [ 80%] (Sampling)
Chain 1 Iteration: 2500 / 3000 [ 83%] (Sampling)
Chain 1 Iteration: 2600 / 3000 [ 86%] (Sampling)
Chain 2 Iteration: 2600 / 3000 [ 86%] (Sampling)
Chain 2 Iteration: 2700 / 3000 [ 90%] (Sampling)
Chain 3 Iteration: 2400 / 3000 [ 80%] (Sampling)
Chain 3 Iteration: 2500 / 3000 [ 83%] (Sampling)
Chain 3 Iteration: 2600 / 3000 [ 86%] (Sampling)
Chain 4 Iteration: 2400 / 3000 [ 80%] (Sampling)
Chain 4 Iteration: 2500 / 3000 [ 83%] (Sampling)
Chain 1 Iteration: 2700 / 3000 [ 90%] (Sampling)
Chain 1 Iteration: 2800 / 3000 [ 93%] (Sampling)
Chain 2 Iteration: 2800 / 3000 [ 93%] (Sampling)
Chain 2 Iteration: 2900 / 3000 [ 96%] (Sampling)
Chain 2 Iteration: 3000 / 3000 [100%] (Sampling)
Chain 3 Iteration: 2700 / 3000 [ 90%] (Sampling)
Chain 3 Iteration: 2800 / 3000 [ 93%] (Sampling)
Chain 4 Iteration: 2600 / 3000 [ 86%] (Sampling)
Chain 4 Iteration: 2700 / 3000 [ 90%] (Sampling)
Chain 4 Iteration: 2800 / 3000 [ 93%] (Sampling)
Chain 2 finished in 1.5 seconds.
Chain 1 Iteration: 2900 / 3000 [ 96%] (Sampling)
Chain 1 Iteration: 3000 / 3000 [100%] (Sampling)
Chain 3 Iteration: 2900 / 3000 [ 96%] (Sampling)
Chain 3 Iteration: 3000 / 3000 [100%] (Sampling)
Chain 4 Iteration: 2900 / 3000 [ 96%] (Sampling)
Chain 4 Iteration: 3000 / 3000 [100%] (Sampling)
Chain 1 finished in 1.6 seconds.
Chain 3 finished in 1.5 seconds.
Chain 4 finished in 1.6 seconds.
All 4 chains finished successfully.
Mean chain execution time: 1.5 seconds.
Total execution time: 1.7 seconds.fit4b_n$summary()# A tibble: 8 × 10
variable mean median sd mad q5 q95 rhat ess_bulk
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 lp__ -170. -169. 1.89 1.73 -173. -167. 1.00 4136.
2 alpha_1[1] 2.53 2.53 0.209 0.204 2.19 2.87 1.00 10825.
3 alpha_1[2] 3.75 3.75 0.217 0.215 3.39 4.11 1.00 10929.
4 sigma_x 1.63 1.62 0.107 0.106 1.46 1.81 1.00 10108.
5 beta_1[1] 0.776 0.768 0.521 0.522 -0.0606 1.64 1.00 6724.
6 beta_1[2] 1.45 1.40 1.17 1.13 -0.390 3.46 1.00 6954.
7 beta_2[1] -0.404 -0.396 0.194 0.191 -0.731 -0.0970 1.00 6755.
8 beta_2[2] -1.69 -1.61 0.666 0.649 -2.92 -0.748 1.00 6854.
# ℹ 1 more variable: ess_tail <dbl>str(sim2_parms)List of 7
$ n : num 116
$ a1 : num [1:2] 2.5 4
$ b1 : num [1:2] 1.7 2.2
$ b2 : num [1:2] -0.67 -1.37
$ sigma_x: num 1.5
$ LoD : num 3
$ latent : logi TRUEfit_summary_n <- fit4b_n$summary() |>
dplyr::select(variable, median, q5, q95) |>
dplyr::filter(variable != "lp__") |>
dplyr::mutate(
truth = c(
#sim2_parms$a0,
sim2_parms$a1[[1]],
sim2_parms$a1[[2]],
sim2_parms$sigma_x,
#sim2_parms$b0,
sim2_parms$b1[[1]],
sim2_parms$b1[[2]],
sim2_parms$b2[[1]],
sim2_parms$b2[[2]]
)
)
pn <-
ggplot(fit_summary_n) +
aes(x = variable, y = median, ymin = q5, ymax = q95) +
geom_hline(yintercept = 0, linetype = "dashed", color = "gray") +
geom_pointrange() +
geom_point(aes(y = truth), shape = 4, color = "red", size = 3, stroke = 1) +
labs(
x = NULL,
y = "Parameter value",
title = "Model-estimated median with 95% CI; x marks true simulation value",
subtitle = "Estimated using censored values without censoring correction"
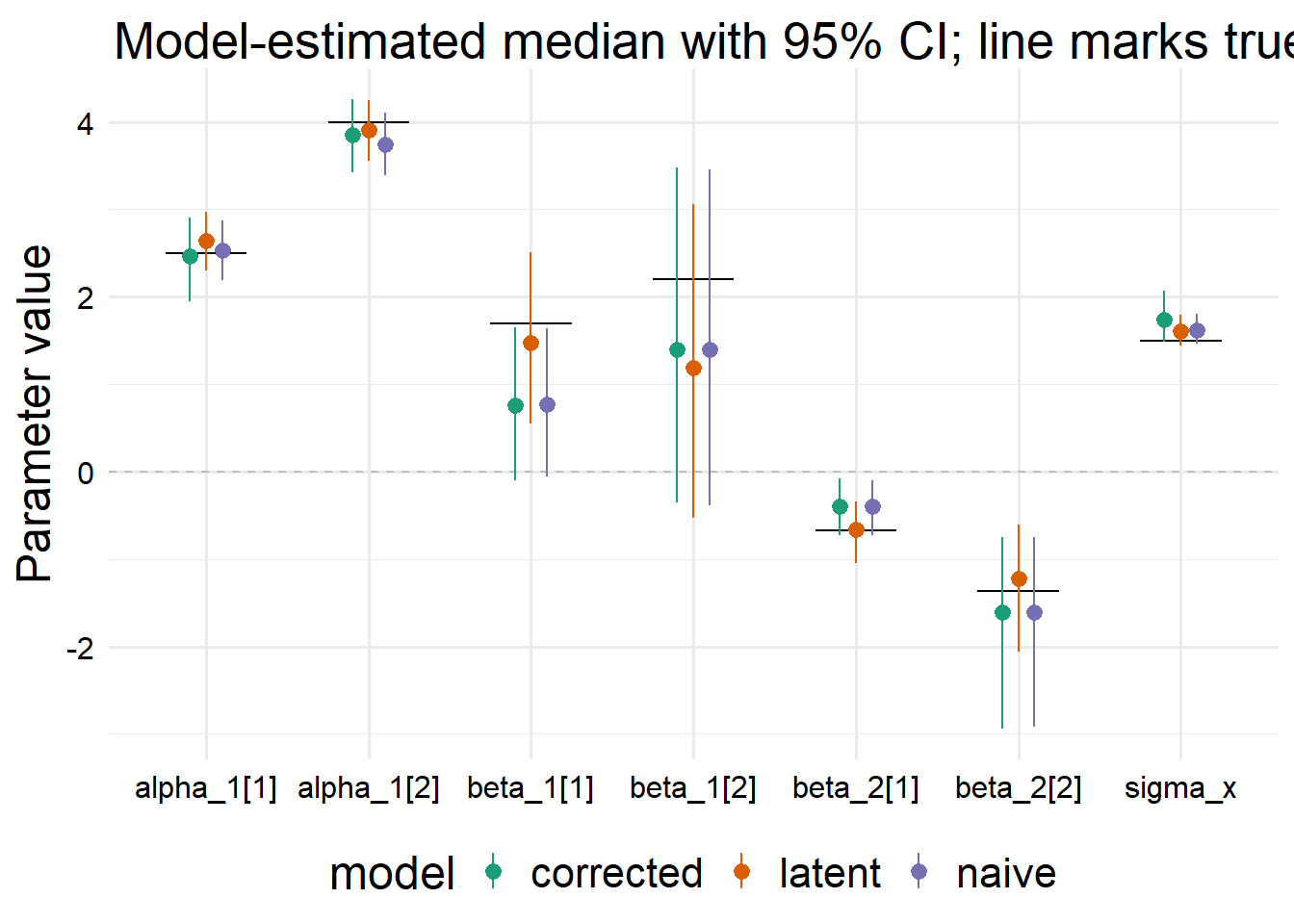
)po / pl / pn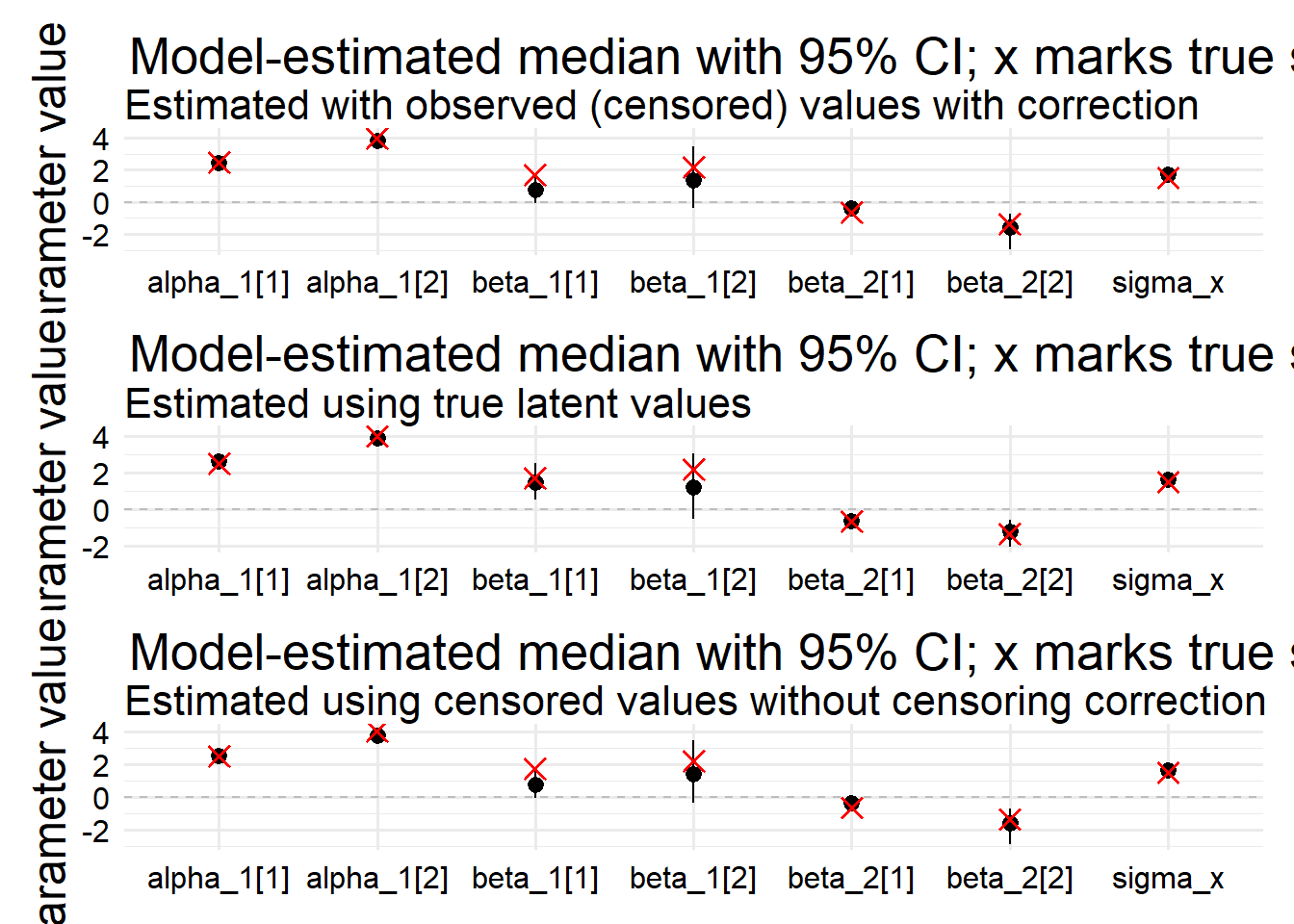
all_fits <-
dplyr::bind_rows(
"corrected" = fit_summary,
"latent" = fit_summary_l,
"naive" = fit_summary_n,
.id = "model"
)
all_fits |>
ggplot() +
aes(
x = variable, y = median, ymin = q5, ymax = q95,
color = model
) +
geom_hline(yintercept = 0, linetype = "dashed", color = "gray") +
geom_crossbar(
aes(y = truth, ymin = truth, ymax = truth),
width = 0.5,
color = "black",
fatten = 0.1
) +
geom_pointrange(position = position_dodge2(width = 0.3)) +
scale_color_brewer(palette = "Dark2") +
labs(
x = NULL,
y = "Parameter value",
title = "Model-estimated median with 95% CI; line marks true value"
)
6.5 Model exploration
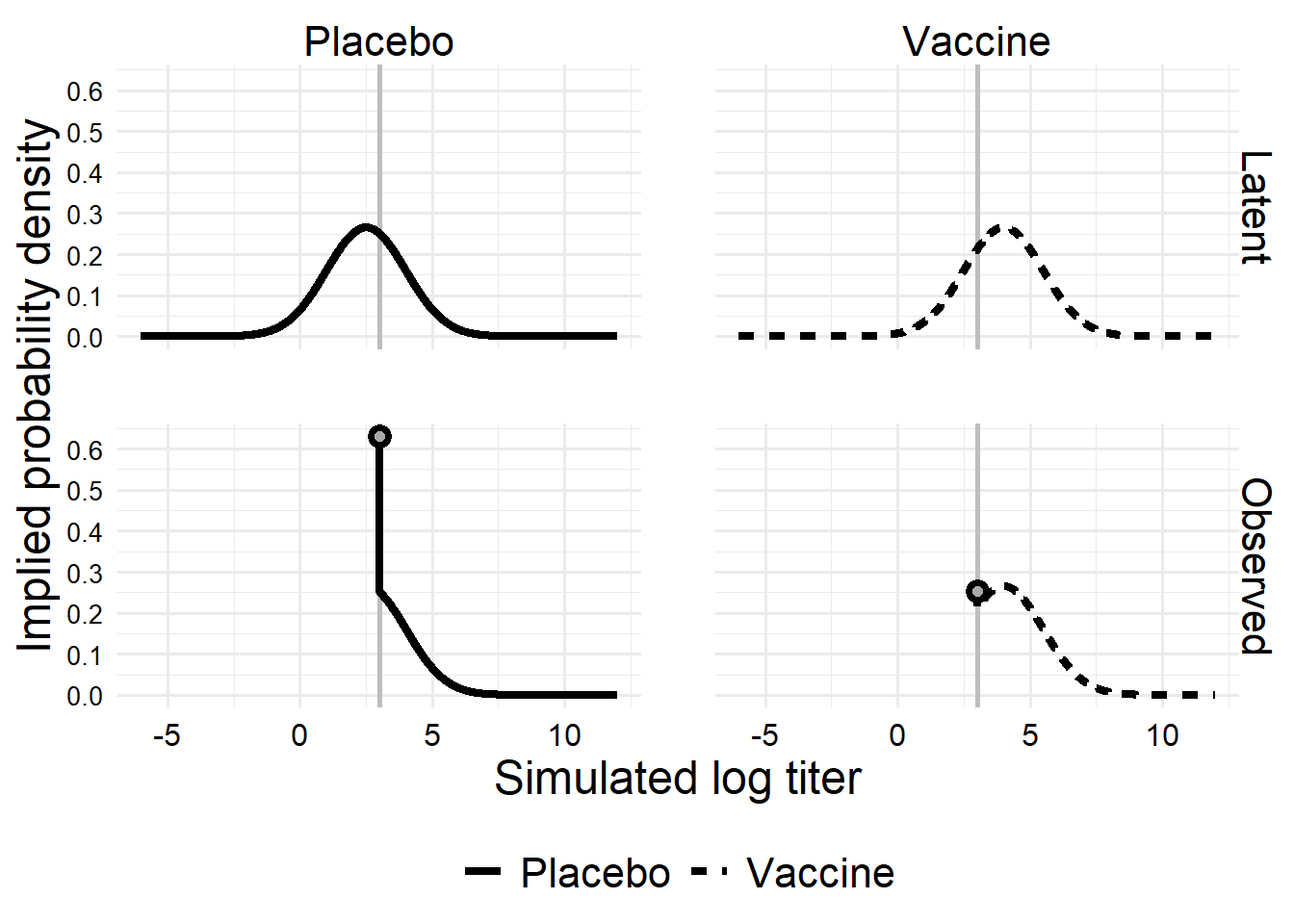
these curves are implied by the model and parameters, not by the simulated data ::: {.cell}
x_vec <- seq(-6, 12, 0.01)
r1 <- sapply(x_vec, \(x) inv_logit(1.5 + 0.2 - 0.67 * x))
r2 <- sapply(x_vec, \(x) inv_logit(1.5 + 0.7 - 1.37 * x))
layout(matrix(c(1, 2, 3, 3), ncol = 2, byrow = TRUE))
plot(x_vec, r2 - r1, ylab = "risk difference", type = "l", xlab = "")
abline(h = 0, lty = 2)
plot(x_vec, r2 / r1, ylab = "risk ratio", type = "l", xlab = "")
abline(h = 1, lty = 2)
lab2 <- latex2exp::TeX(r"($Pr(y_{i} = 1 \ | \ x_{i}, T_{i})$)")
plot(
NULL, NULL,
ylim = c(0, 1),
xlim = c(-6, 12),
yaxs = "i",
xaxs = "i",
xlab = "Simulated log titer",
ylab = lab2
)
lines(x_vec, r1, lty = 2, lwd = 1.5) # placebo
lines(x_vec, r2, lty = 1, lwd = 1.5) # vaccine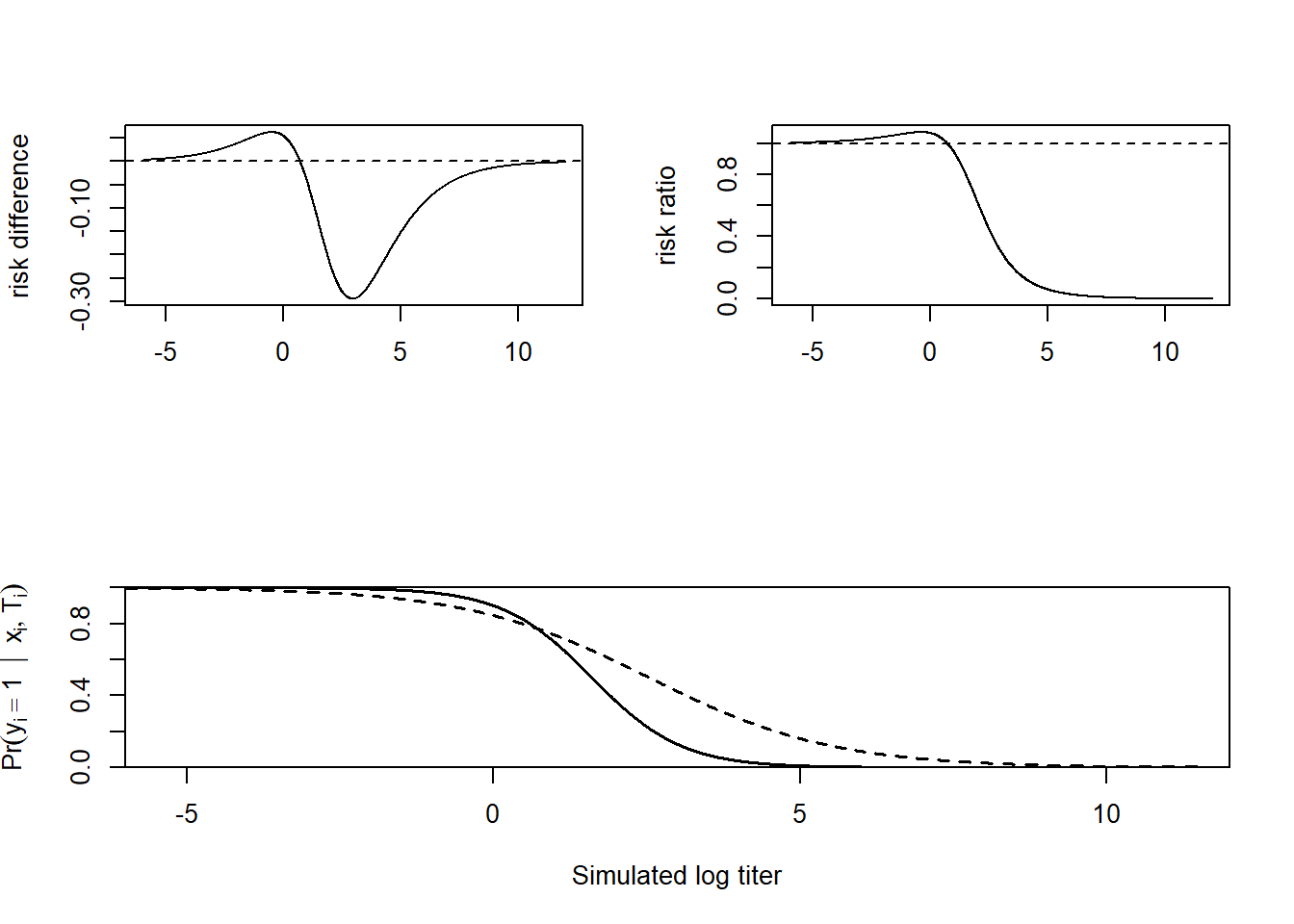
# IDK what's wrong with the legend, seems it doesn't like layout.
# switch to ggplot to fix
#legend(x = 9, y = 0.8, c('Unexposed', 'Exposed'), lty = c(2, 1), lwd = 2):::
x_dens <-
tibble::tibble(
Latent = x_vec,
Observed = dplyr::if_else(
x_vec < sim2_parms$LoD,
sim2_parms$LoD,
x_vec
),
Placebo = sim2_parms$a1[1],
Vaccine = sim2_parms$a1[2]
) |>
tidyr::pivot_longer(
cols = c(Placebo, Vaccine),
names_to = "t",
values_to = "mu"
) |>
tidyr::pivot_longer(
cols = c(Latent, Observed),
names_to = "o",
values_to = "x"
) |>
dplyr::mutate(
d = dplyr::if_else(
o == "Latent",
dnorm(x, mean = mu, sd = sim2_parms$sigma_x),
crch::dcnorm(
x, mean = mu, sd = sim2_parms$sigma_x,
left = sim2_parms$LoD, right = Inf
)
)
)
anno_df <-
x_dens[1:4, ] |>
dplyr::filter(o == "Observed")
x_dens |>
ggplot() +
aes(x = x, y = d, linetype = t, group = t) +
geom_vline(
xintercept = sim2_parms$LoD,
linetype = 1, linewidth = 1, color = "gray"
) +
geom_line(linewidth = 1.5) +
geom_point(
data = anno_df,
size = 2,
stroke = 2,
shape = 21,
color = "black",
fill = "darkgray"
) +
facet_grid(vars(o), vars(t)) +
scale_linetype_discrete(name = NULL) +
scale_x_continuous(breaks = scales::breaks_pretty()) +
scale_y_continuous(breaks = scales::breaks_pretty()) +
labs(
x = "Simulated log titer",
y = "Implied probability density"
) +
#coord_cartesian(expand = FALSE, ylim = c(-0.01, 0.28)) +
theme(axis.text.y = element_text(size = 10))